Introduction
Probiotics are described by FAO/WHO as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Most probiotic strains that are beneficial for human health are included in the “Lactic acid bacteria” (LAB) group. However, the biological activities of probiotic microorganisms that are beneficial to the host may vary depending on the species or even the strain, the number of probiotic cells used per day, and the physiological characteristics of the host (Hawrelak, 2003; Salminen and Gueimonde, 2004; Tannis, 2008).
LABs have long been used as starter cultures for foods and beverages as they can improve the nutritional value, organoleptic properties and shelf life of fermented food (Florou-Paneri et al., 2013). The production of various foodstuffs with the lactic acid fermentation process is one of the oldest inventions of humanity. Many different societies in various parts of the world have used lactic acid fermentation to keep foodstuffs, such as milk, meat, vegetables and fish, without deterioration. Fermented foods and beverages were previously produced by spontaneous fermentation (self-fermentation) with microbiota found naturally in raw materials. Subsequently, the food industry opted for selected starter cultures to be added directly to the food. Thus, the lactic acid fermentation process could be controlled at an optimum level and standardization of the final product was achieved (Bintsis, 2018).
Various products that are traditionally fermented are produced without the use of a starter culture are rich resources for the isolation of LAB strains with probiotic potential. Moreover, since these have long been a part of the human diet, the isolated LABs and some of their metabolites, called postbiotics, are considered safe for human health (Zielińska and Kolożyn-Krajewska, 2018). However, the LAB content of many traditionally fermented food varieties and their probiotic potentials are still unknown. Therefore, studies to obtain new probiotic bacterial strains from traditional foods have recently gained importance (Chander et al., 2018).
Turkey is a very rich country in terms of the fermented product diversity. It has been reported that 160–193 types of cheese are produced in different regions of Turkey (Çetinkaya, 2005; Swan, 2005). Kargi tulum cheese is one of the artisanal cheeses produced by traditional methods in Turkey (Çetinkaya, 2005). This cheese is produced by small-scale dairies using raw whole sheep milk, goat milk, cow milk, buffalo milk or a mixture of these. Afterwards, the curds are compressed into goat-skin bags to ensure that no entrapped air remains. Goat-skin bags are the material traditionally used to package tulum cheese. After this preparation, the Kargi tulum cheese is then incubated for at least 3 months (at 4°C to 6°C) in the uplands (personal communication with local producer). Kunduhoglu et al. (2012) previously reported that the LAB strains were responsible for the spontaneous fermentation of Kargi tulum cheese. However, many beneficial properties and characteristics of the LAB species found in the Kargi tulum cheese remained unknown. Therefore, this study aimed to evaluate the in vitro probiotic characteristics of the L. brevis KT38-3 strain isolated from artisanal Kargi tulum cheese, and the strain was identified by 16S rRNA gene sequencing.
Materials and Methods
In this study, all tests were at least triplicated except for the genotypic species identification. All incubations in this study were performed for 24 h at 37°C (aerobically) unless otherwise specified.
The KT38-3 strain used in this study was isolated in our previous study. First, the essential probiotic characteristics of this strain were determined, and then species level identification was done by 16S rRNA sequencing. The genomic DNA of the KT38-3 strain was isolated with GeneMATRIX Bacterial & Yeast DNA isolation kit (EurX, Gdańsk, Poland) according to the supplier’s specifications. The thermo Scientific Nanodrop 2000 (Massachusetts, MA, USA) device was used to determine the DNA quantity. The targeted gene regions were amplified using the 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-TACGGYTACCTTGTTACGACTT-3') universal 16S rRNA bacterial primers. The PCR procedure was carried out with the same method as Kunduhoglu and Hacioglu (2021). The sequences obtained were compared with GenBank database using the BLAST.
L. brevis KT38-3 was inoculated on MRS broth (Sigma-Aldrich, St. Louis, MO, USA), and after aerobic incubation, the culture was centrifuged at 4,400×g for 10 min at 4°C (Sigma-Aldrich, 3–16 K). Then the supernatant and cell pellet were separated. The cell free supernatant of the L. brevis KT38-3 culture (namely KT38-3-CFS) was sterilized with a 0.22 μm pore size filter. The cell pellet was used for preparing the L. brevis KT38-3 cell suspension (namely KT38-3-CS). The cell pellet was washed twice with phosphate-buffered saline (PBS, pH=7.2) by centrifugation, then the cell density was adjusted (108 CFU/mL) according to the 0.5 McFarland standard.
Concentrated HCl or 5 M NaOH were used to adjust the MRS broth pH to 2, 3, 4, 5, 6, or 7. Each tube of this series was inoculated with 0.1 mL KT38-3-CS. All tubes were incubated at 37°C for 4 h. During the incubation, 0.1 mL aliquots were taken from each tube at one-hour intervals. These samples were spread on the surface of MRS agar (Sigma-Aldrich) plates and the colony numbers (CFU/mL) were determined after the incubation.
The pH of the MRS medium brand we use was 5.8. Therefore, L. brevis KT38-3 cultured in MRS broth (pH=5.8) was the positive control in all of the survival tests.
First, the Ox-bile concentration of the MRS broth tubes was set at 0.3%, 0.5%, or 1.0% (w/v); Sigma-Aldrich). Then, 0.1 mL KT38-3-CS was added to each tube and incubated for 6 h at 37°C. A 0.1 mL sample was taken from each tube at one-hour intervals and colony growth was checked as described above.
The final pH of the artificial gastric juice (NaCl 0.85% w/v and pepsin 0.5% w/v) was adjusted to 2.0 with 1 M HCl. A total of 90 mL artificial gastric juice was inoculated with 10 mL of KT38-3-CS. After mixing, the initial bacterial count was determined by colony count and incubated at 37°C for 4 h with shaking. The final bacterial number was determined after the incubation. This artificial gastric juice culture was used for the intestinal juice test. For this, KT38-3 cells and artificial gastric juice were separated by centrifugation at 4,400×g for 10 min. The pellets were washed with PBS by centrifugation at 4,400×g for 10 min. Then, the pellets were resuspended in artificial intestinal fluid (pancreatin 0.1% w/v, bovine bile salts 0.15% w/v; pH=7.0). A total of 90 mL of artificial intestinal fluid was inoculated with 10 mL of the KT38-3-CS obtained from the artificial gastric juice. After mixing, the initial bacterial count was determined by colony count and incubated at 37°C for 4 h with shaking. The initial and final bacterial numbers were described above (Zárate et al., 2000; Grimoud et al., 2010).
The cell surface hydrophobicity of L. brevis KT38-3 was assessed by measuring the level of microbial adhesion to hydrocarbons (Kotzamanidis et al., 2010). KT38-3-CS was resuspended in 3 mL of 0.1 M KNO3, and the cell suspension optical density (OD600) was measured (A0). Then Ksilene (1 mL) was added to this cell suspension, vortexed for 2 min, and incubated at 37°C, for 1 h. Then, the water and xylene phases were separated, and the optical density of the aqueous phase was measured (A1). The hydrophobicity of L. brevis KT38-3 was classified according to Rosenberg and Gutnick (1980). The percentage of the cell surface hydrophobicity was calculated using the following formula:
KT38-3-CS was vortexed for 30 s and incubated for 4 h at 37°C. The OD600 of KT38-3-CS was measured before (A0) and after (At) incubation using a spectrophotometer. The autoaggregation percentage was calculated as follows (Abdulla et al., 2014):
KT38-3-CS (1 mL) and the cell suspension of Escherichia coli O157:H7 (1 mL) were mixed and incubated at 37°C without agitation. The absorbances of this mixture were measured at different times (0 or 4 h; at 600 nm). In addition, absorbance was determined for each bacterial suspension alone. The coaggregation percentage was calculated as follows:
where AE. coli and AL. brevis represent the A600nm of the separate bacterial suspensions in control tubes, and Amix represents the absorbance of the mixed bacterial suspension at 4 h.
A deoxyribonuclease (DNase) test was performed to determine the ability of L. brevis KT38-3 to hydrolyze DNA. DNase agar plates (Sigma-Aldrich) were inoculated with KT38-3-CS. After incubation, 1 N HCl was poured into the medium. If a clear zone occurred surrounding the colony, it was considered a positive result (Ejiofor et al., 2018).
The KT38-3-CS was seeded on sheep blood agar plates. After incubation, if there was a greenish or clear zone surrounding the colony, it was considered as α-hemolysis or β-hemolysis, respectively (Pavlov et al., 2004).
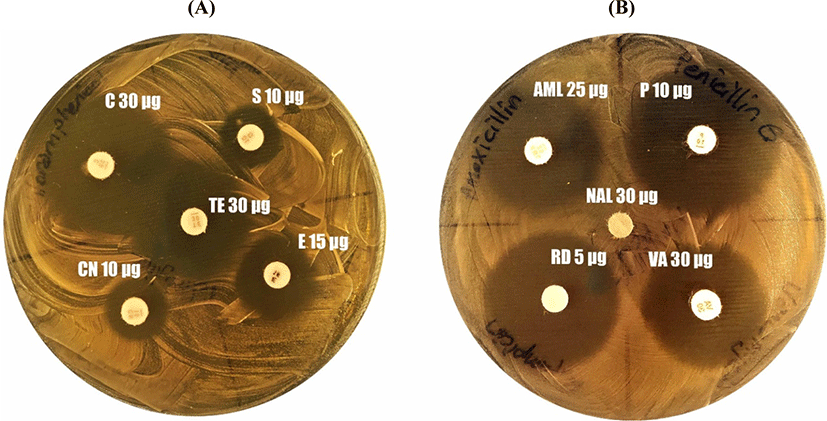
The inhibitory activity of several of the antibiotics listed in (Fig. 1) against L. brevis KT38-3 were determined. The L. brevis KT38-3 was inoculated in MRS broth and incubated overnight at 37°C. The fresh MRS broth culture of L. brevis KT38-3 (the final concentration was 108 CFU/mL; 0.1 mL) was poured and spread onto MRS agar plates. Then antibiotic discs were placed on the surface of MRS agar plates. The inhibition zone diameters were measured after incubation. Then the sensitivity/resistance of the strain was evaluated according to the CLSI breakpoints (2017).
The L. brevis KT38-3 enzymatic activity spectrum was determined with API-ZYM (BioMérieux, Lyon, France) test kits. The types of enzyme activity that were tested are listed in Table 1. Each cupule of the API ZYM was inoculated with KT38-3-CS. After 4 h of incubation at 37°C, the results of the 19 enzyme (Table 1) activities tested for L. brevis KT38-3 were evaluated according to the manufacturer's instructions.
The ability of L. brevis KT38-3 to ferment carbohydrates was determined by API CH50 test kits (BioMérieux). The names of the 49 carbohydrates tested are given in Table 2. Each cupule of the API CH50 was inoculated with KT38-3-CS, and the test results were evaluated according to the manufacturer’s instructions and API-WEB.
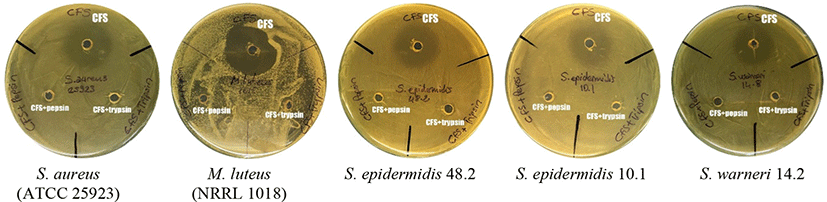
The antagonistic activity of the cell free supernatant (KT38-3-CFS) of L. brevis KT38-3 was determined with an agar well diffusion assay. L. brevis KT38-3 could produce hydrogen peroxide (data not shown). Therefore, catalase (1 mg/mL; Sigma-Aldrich) was added to KT38-3-CFS to exclude the antimicrobial effect of hydrogen peroxide. In addition, the pH of the KT38-3-CFS was adjusted to 6.5 (with 5 M NaOH) to exclude the antimicrobial effects of the organic acids produced by L. brevis KT38-3. Then KT38-3-CFS was filter sterilized. Catalase-free KT38-3-CFS (pH=6.5) served as the control. The indicator test bacteria including the methicillin and vancomycin-resistant (M&V-R), methicillin-resistant (M-R) and vancomycin-resistant (V-R) strains that were used in this assay are listed in Table 3. Briefly, the fresh Brain Heart Infusion broth cultures of indicator test bacteria (the final concentration was 108 CFU/mL) were poured and spread onto Brain Heart Infusion agar plates, the wells were bored into the agar using a sterile cork borer (6 mm in diameter). Then the 0.1 mL filter sterilized KT38-3-CFS was added to the wells. After incubation at 37°C for 24 h, the inhibition zone diameter surrounding the well was measured (Abanoz and Kunduhoglu, 2018). In addition, the antimicrobial effects of KT38-3-CFS (catalase-free, pH=6.5) treated with pepsin (1 mg/mL) or trypsin (1 mg/mL) were determined to show that the antimicrobial effect is due to protein-like compounds. The antimicrobial effects of KT38-3-CFS treated with pepsin or trypsin were tested on KT38-3-CFS-susceptible strains.
The antioxidant capacity of the CFS of L. brevis KT38-3 (KT38-3-CFS) was determined with the DPPH (1,1-diphenyl-2-picrylhydrazyl) method (Brand-Williams et al., 1995). DPPH was dissolved in methanol (60 μM). In the dark, 0.1 mL aliquots of the KT38-3-CFS were added to test tubes containing 2.9 mL of the freshly prepared DPPH radical solution, and it homogenized by shaking and incubated for 30 min. Methanol served as the blank control, and the uninoculated MRS broth served as the blank. Butylated hydroxytoluene (BHT) and ascorbic acid (100 μg/mL) were used as positive controls. The absorbance was measured at 517 nm. The radical scavenging ability of KT38-3-CFS was calculated by using the following formula:
where Asample is the absorbance of KT38-3-CFS and Acontrol is the absorbance of the blank.
The antioxidant activity of KT38-3-CFS was also determined with the ABTS (2,2'-Azino-bis[3-ethylbenzothiazoline-6-sulfonic]) radical cation decolorization assay (Re et al., 1999).
The ABTS radical cation (ABTS·+) was prepared by mixing ABTS (7 mM) with potassium persulfate (2.45 mM), (1:1, v/v). Following the addition of 0.1 mL of the KT38-3-CFS (sample) to 0.9 mL of the ABTS·+ solution, absorbance was measured after 1 min. The absorbance was measured at 734 nm in this assay. The positive controls and blank series were set as in the DPPH assay. The radical cation scavenging ability of KT38-3-CFS was calculated by following formula (Tripathi et al., 2007):
where Asample is the absorbance of KT38-3-CFS, Acontrol is the absorbance of the blank after a reaction time of 1 min.
Results and Discussion
In this study, the in vitro probiotic properties of the KT38-3 strain isolated from artisanal Kargi tulum cheese were investigated, and this strain was then identified by the 16S rRNA gene sequence. The sequences obtained from the KT38-3 strain were compared to the GenBank database using BLAST. The 16S rRNA sequences of the KT38-3 strain were 100% similar to the 16S rRNA sequences of the L. brevis MN049503.1 registered in the GenBank database. The sequence of L. brevis KT38 is available from NCBI under the accession number MN334210.
Some of the L. brevis strains are known for their probiotic potential, and a few of them are employed as starter cultures (Annuk et al., 2003). L. brevis is a common LAB species found in the microbiota of fermented cheeses (Guley et al., 2014/15; Mugampoza et al., 2020; Skelin et al., 2012). The isolation of the L. brevis KT38-3 strain from artisanal Kargi Tulum cheese is in accordance with the data in the literature. Also, many strains of this species are used for biotechnological purposes (Kaur et al., 2018; Yue et al., 2013; Zhang et al., 2012). Due to being a member of the microbiota of various traditionally fermented foods, L. brevis has a Generally Recognized as Safe (GRAS) status (Singh et al., 2019).
The bacteria which is to be used as probiotics must be resistant to the harsh conditions of the human digestive system. The pH in the gastrointestinal region of healthy individuals varies between 2 and 6.7 (Fallingborg, 1999). The physiological bile salt concentration in the small intestine of a healthy individual is 0.3%. Therefore, the first feature to be tested in probiotic bacteria selection should be the resistance to gastric acidity and bile salts. In our study, the survival of L. brevis KT38-3 at pH 2 was very close to that of the control at pH 5.8 (Table 4). In addition, the survival of L. brevis KT38-3 at pH=3–7 was equal to that of the control series. Moreover, L. brevis KT38-3 maintained its stability over 6 h at all the bile salt concentrations tested (0.3%, 0.5%, and 1%) (Table 4). Bile salts secreted in the small intestine can damage the cell membranes of probiotic bacteria by hydrolyzing the lipids and fatty acids (Pennacchia et al., 2004). Thus, probiotic bacteria must be tolerant of bile salt. In a similar study, the survival percentage of 5 strains of L. brevis ranged from 33% to 64% when at pH 2.5 for 4 h. They found that the bile salt tolerance (0.3% for 4 h) of their 5 isolates was from 94% to 99% (Tokatlı et al., 2015). Kariyawasam et al. (2020) determined that the survival percentage of the L. brevis KU200019 strain at pH=2.5 and 0.3% bile was 99.4% and 115.1%, respectively. However, Fang et al. (2018) found that the L. brevis BBE-Y52 strain lost its viability 3.6 log unit after 3 h at pH 2. The difference in the pH results is an important indicator that the probiotic properties change depending on the strain.
It was determined that the number of L. brevis KT38-3 cells surviving in the artificial gastric juice was almost equal (93%) to the number of cells in the control group at the end of 4 h (Table 4). In addition, the L. brevis KT38-3 cells that survived in artificial gastric juice were transferred into the artificial intestinal fluid, then the percentage of the cells that survived after 6 h was determined. As a result, we determined that 77% of the L. brevis KT38-3 cells inoculated in the artificial intestinal fluid survived after 4 h (Table 4). In a similar study, the L. brevis CCMA1284 strain was reported to survive in artificial gastric juices at pH 2.0 (for 4 h) and artificial intestinal fluid at pH 7 (4 h) (Fonseca et al., 2021). Tokatlı et al. (2015) reported that the viability percentages of the five L. brevis strains in artificial gastric and intestinal fluid were 76%–99% and 42%–63%, respectively. Vecchione et al. (2018) demonstrated the survival of ten commercial probiotic formulations in artificial gastric juice (pH=1.5, for 120 min) and intestinal fluid (pH=8.0, for 360 min). As a result, they reported that the number of cells in the VSL3, Yovis and Enterogermina preparations did not change in artificial gastric fluid after 120 min, but the cell number of the other probiotic formulations decreased after the 30–120 min incubation period. While the cell number of the Enterogermina preparations in simulated intestinal fluid increased at the end of 360 min, the number of the VSL3 and Yovis formulations decreased after the 240–360 min incubation period. These in vitro survival tests showed that it is possible for L. brevis KT38-3 to survive the harsh conditions of the human digestive system.
The cell surface hydrophobicity and autoaggregation capability are the basic parameters that show the capacity of a probiotic candidate bacterium to adhere to the intestinal epithelium of the host and form biofilms (Abdulla et al., 2014; Tareb et al., 2013). In our study, the percentage of the cell surface hydrophobicity of L. brevis KT38-3 was 85.0%, and its autoaggregation capacity was 59.4% (Table 4). Fonseca et al. (2021) determined that the autoaggregation percentage of the L. brevis CCMA 1284 was 22.09%. Somashekaraiah et al. (2019) reported that the hydrophobicity and autoaggregation capacity of the L. brevis MYSN 106 strain were 77.82% and 75.95%, respectively. Meira et al. (2012) reported the hydrophobicity level of the L. brevis SM-A and L. brevis SM-B strains as 88.0% and 34.6%, respectively. They determined the autoaggregation ability of the L. brevis SM-A and L. brevis SM-B strains as 45.2% and 41.9%, respectively. The hydrophobicity of the four L. brevis strains was determined by Vasiee et al. (2018), and it ranged from 28%–76%.
The ability of probiotic cells to co-aggregate with pathogenic cells is a feature that increases their probiotic usefulness. Campana et al. (2017) reported a correlation between the autoaggregation and coaggregation properties of their strains. In our study, L. brevis KT38-3 also showed good coaggregation and autoaggregation ability. The coaggregation percentage of L. brevis KT38-3 with E. coli O157:7 was 47.4% (Table 4). Kariyawasam et al. (2020) reported that the coaggregation percentage for L. brevis KU200019 and E. coli O157:H4 FRIK 125 was 32.7%. Fonseca et al. (2021) determined that the coaggregation percentage of L. brevis CCMA 1284 with the entoropathogenic E. coli was 2.39%. Ramos et al. (2013) reported that the coaggregation capacities of L. brevis FFC199 and L. brevis SAU105 with E. coli JM109 were 1.1% and 28.1%, respectively. The different percentages of hydrophobicity, autoaggregation and coaggregation that were obtained from different L. brevis strains showed, once again, that probiotic characteristics are strain specific.
Bacteriocins are antimicrobial peptides ribosomally synthesized by bacteria. The bacteriocin production characteristic is an important criterion in the selection of probiotic strain candidates (Dobson et al., 2012). Many studies have reported the antagonistic effect of bacteriocins on the bacteria that cause various systemic urogenital, gastrointestinal, respiratory and skin infections, including bacteria with multiple antibiotic resistance (Dicks et al., 2011). Therefore, the inhibitory effect has been tested for KT38-3-CFS against various indicator test bacteria (Table 3). The LAB produces organic acids that have an antimicrobial effect and postbiotics, such as H2O2, in the environment in which they grow. For this reason, by adding catalase enzyme to CFS and neutralizing the pH, the inhibitory effect of H2O2 and organic acids became negligible. As a result, the CFS of L. brevis KT38-3 showed an antimicrobial effect against S. aureus (ATCC 25923) and M. luteus (NRRL 1018). In addition, CFS showed an inhibitory effect against S. epidermidis 10.1 (M-R), S. epidermidis 48.2 (M&V-R) and S. warneri 14.2 (M&V-R). The antimicrobial activity of KT38-3-CFS was lost after treatment with pepsin and trypsin enzymes (Fig. 2). This finding suggests that the antimicrobial effect is due to a protein/peptide substance(s). In our previous study, the supernatant obtained from the L. brevis KT16-2 strain was found to have a broader antimicrobial activity spectrum, including activity against M-R S. epidermidis and M&V-R S. warneri (Kunduhoglu and Hacioglu, 2021). Silva et al. (2019) determined that the L. brevis A6, B16 and E35 strains produce bacteriocin-like antimicrobial peptides against Escherichia coli, Enterococcus faecalis, L. monocytogenes, Salmonella Tyhimurium and Shigella flexneri. However, Fonseca et al. (2021) reported that the L. brevis CCMA 1284 strain had no antimicrobial effect on any indicator test bacteria such as E. coli CDC 055, B. cereus ATCC 14579, S. aureus ATCC 5674 and L. monocytogenes ATCC 19117.
The antioxidant potential of the CFS obtained from L. brevis KT38-3 was tested by the DPPH and ABTS methods. In our study, DPPH radical scavenging capacity was calculated as 54.9%. The ABTS scavenging activity of L. brevis KT38-3 was 48.7% (Table 4). In our previous study, L. brevis KT16-2 had a DPPH scavenging activity of 71.0% and an ABTS scavenging activity of 54.1% (Kunduhoglu and Hacioglu, 2021). Jang et al. (2019) reported that the DPPH scavenging activity of the L. brevis KU15153 strain was 44.1%. Somashekaraiah et al. (2019) determined that the hydroxyl radical scavenging activity of their L. brevis strain was 77.9%. In another study, the linoleic acid inhibition percentage of the L. brevis SM-A and L. brevis SM-B strains were 26% and 32%, respectively (Meira et al., 2012). Strain specific factors such as the bacterial cell wall composition, enzyme activity and metabolite production have effects on the antioxidant activity (Jang et al., 2019). Considering the results, L. brevis KT38-3, which shows antioxidant activity in vitro, is promising as a probiotic. However, its antioxidant activity should also be supported by in vivo experiments.
Bacteria that are used as probiotics are expected to not produce enzymes such as α-chymotrypsin, β-glucuronidase or N-acetyl-β-glucosaminidase, which are associated with intestinal diseases (de Melo Pereira et al., 2018). The increases in β-glucuronidase levels in feces is thought to be associated with gastric cancer and inflammatory bowel disease (Jang et al., 2019). API ZYM tests showed that the enzyme spectrum of L. brevis KT38-3 was quite broad (Table 1). L. brevis KT38-3 had a high level of β-galactosidase activity. This means that L. brevis KT38-3 could be beneficial for individuals with lactose intolerance. Additionally, L. brevis KT38-3 did not produce α-chymotrypsin, β-glucuronidase, or N-acetyl-β-glucosaminidase. Similar to our study, it was determined that the L. brevis S82 strain had high aminopeptidase, β-galactosidase and β-glucosidase enzyme activity but not β-glucuronidase enzyme activity (Abouloifa et al., 2020). In addition, Kariyawasam et al. (2020) reported that L. brevis KU200019 and L. brevis ATCC 148691 did not show the enzymatic activities associated with intestinal diseases.
The carbohydrate fermentation profile of L. brevis KT38-3 was determined with an API 50CHL test kit (Table 2). L. brevis KT38-3 was able to ferment 26 carbohydrates, including low-digestible carbohydrates such as mannitol, sorbitol and inulin (Grabitske and Slavin, 2009). The ability of L. brevis KT38-3 to ferment lactose and inulin increases its probiotic potential. Similar to our results, Meira et al. (2012) determined that the L. brevis SM-A and L. brevis SM-B strains ferment lactose, and their β-galactosidase enzyme activities were 600 and 300 Miller Units, respectively.
Microorganisms that are used as probiotics should not be virulent. For this reason, we examined whether L. brevis KT38-3 produces virulence-related enzymes such as DNase and hemolysin (Table 4). In vitro test results showed that this strain has no DNase or hemolytic activity. In similar studies, it has been reported that the L. brevis strains do not produce hemolysis and DNase enzymes (Abouloifa et al., 2020; Hasali et al., 2018; Pavli et al., 2016; Somashekaraiah et al., 2019). However, these phenotypic tests do not indicate the absence of the transferable virulence genes in the genome of a bacterium. Therefore, these results should be supported by further genotypic assays.
Probiotic bacteria should not carry antibiotic resistance genes that are transferred horizontally because antibiotic resistance genes can be transferred to bacteria in the gastrointestinal microbiota. Therefore, we wanted to determine the sensitivity of L. brevis KT38-3 to some therapeutic antibiotics. The sensitivity of L. brevis KT38-3 to some therapeutic antibiotics, according to the breakpoints of CLSI (2017); susceptible (S), moderate (I), or resistant (R). As a result, it was determined that L. brevis KT38-3 was sensitive to all tested antibiotics except streptomycin (I), erythromycin (I), gentamicin (I) and nalidixic acid (R) (Fig. 1). Results from studies on the antibiotic sensitivity of the different L. brevis strains showed that the sensitivity was strain-specific (Fang et al., 2018; Fonseca et al., 2021; Jamaly et al., 2011; Pavli et al., 2016; Silva et al., 2019; Somashekaraiah et al., 2019). However, it must be shown genotypically whether they carry transferable antibiotic resistance genes or not.
Conclusion
It is difficult to find a probiotic strain with all the desired properties. However, our L. brevis KT38-3 strain has many of the essential probiotic characteristics. This strain is able to survive in low pH levels, bile salts, and artificial gastric and intestinal juice. Also, the bacteriocin-like substance/s produced by L. brevis KT38-3 showed antimicrobial activity against some pathogens including the methicillin and/or vancomycin-resistant strains that were used. L. brevis KT38-3 had an antioxidant effect, autoaggregation, coaggregation and capacity for hydrophobicity. It was also sensitive to most of antibiotics used in this study. Moreover, it did not produce DNase and the hemolysin enzymes, but it did have β-Galactosidase activity that ferment lactose. Therefore, considering these in vitro probiotic properties, L. brevis KT38-3 is a promising strain and has the potential to be used as a probiotic supplement. Supporting these features with in vivo experiments to evaluate their potential health benefits will be the subject of our future work.













