Introduction
Fermentation is one of the oldest known food preservation techniques and has been used since ancient times to produce various fermented meat products to protect the meat. Fermented meat products are important sources of valuable nutrients such as protein, fat, essential amino acids, minerals and vitamins (Ojha et al., 2015). Sucuk is a traditional Turkish dry-fermented sausage that is one of the most popular meat products produced in Turkey. In addition, it is popular in many Middle Asians, Middle Eastern, Southeastern European and Northern European countries (Ercoşkun and Özkal, 2011). Sucuk is produced from a mixture of beef, sheep and/or water buffalo meats, beef fat and/or sheep tail fat, salt, sugar, nitrite/nitrate and various spices. In the traditional Sucuk production process, the prepared Sucuk batter is stuffed into special casings, air-dried in a bovine small intestine and then fermented and ripened to develop its typical sensory characteristics (Ercoşkun and Özkal, 2011; Kaban, 2013).
Enterococcus is a large genus of lactic acid bacteria (LAB) that is important both in food and clinical microbiology (Yogurtcu and Tuncer, 2013). Some species of this genus, especially E. faecalis and E. faecium, are the relevant components of the bacterial population of some traditional cheeses (Cariolato et al., 2008; Yogurtcu and Tuncer, 2013) and sausages (Landeta et al., 2013; Yüceer and Özden Tuncer, 2015) produced in different European countries. On the other hand, some enterococci are considered as opportunistic human pathogens that often cause hospital-acquired infections such as endocarditis, bacteremia and urinary tract infection. Enterococcal infections are predominantly associated with E. faecalis and E. faecium (Ogier and Serror, 2008). Although antibiotic resistance is not in itself a virulence factor, multiple antibiotic resistance of enterococci is a contributing factor to their pathogenesis. Enterococci have intrinsic and acquired antibiotic resistance, which is encoded on the chromosome and plasmids or transposons, respectively (Beceiro et al., 2013; Yogurtcu and Tuncer, 2013). The acquired antibiotic resistance genes can be horizontally transferred by mobile genetic elements from other strains, either distantly or closely related. In recent years, attention to the presence of antibiotic resistance in non-pathogenic bacteria has increased because they may act as reservoirs for antibiotic resistance genes (Talon and Leroy, 2011). The human gastrointestinal tract provides an ideal combination of factors for antibiotic resistance genes to arise and spread through bacterial populations (Huddleston, 2014).
Limited data are available on the antibiotic resistance of enterococci isolated from Sucuk and the presence of their antibiotic resistance genes. The objective of the present investigation was to isolate enterococci in Sucuk produced without using starter culture and to analyze isolates for their biodiversity, antibiotic resistance patterns and the presence of some antibiotic resistance genes.
Materials and Method
A total of 20 Sucuk samples produced without using a starter culture were obtained from 20 different local manufacturers in Afyonkarahisar province, Turkey. Sucuk samples were purchased randomly between May and September 2013. Each 25 g Sucuk sample was homogenized with 225 mL sterile physiological water (0.85% NaCl, w/v) in a Waring blender (8011 ES HGB2WTS3, USA). 100 µL of serial decimal dilutions from homogenized Sucuk samples were inoculated onto Kanamycin Aesculin Azide (KAA) agar (LAB M Ltd., UK) and incubated at 37°C for 24-48 h. Typical enterococcal colonies were randomly selected from KAA agar and transferred into de Man, Rogosa and Sharpe (MRS) broth (LAB M). Stock cultures were stored at −20°C in MRS broth with 20% glycerol.
Total DNAs of presumptive enterococci were extracted from overnight cultures, and grown in MRS broth at 37°C, as previously described by Cancilla et al. (1992).
The isolates were identified at the genus level, using Gram staining, catalase and cultural tests such as, growth in MRS broth at 10°C, 45°C, pH 9.6 and 6.5% NaCl (w/v). In addition, isolates were tested for resistance to heat at 60°C for 30 min (Morandi et al., 2006). Identification of isolates at species level was done by 16S rDNA sequencing with pA and pE' universal bacterial primers (Edwards et al., 1989). Polymerase chain reaction (PCR) was performed in 50 μL reaction mixtures, using 3 μL of DNA solution, 1 μL of each primer, 20 μL nuclease-free water and 25 μL PCR master mix (Fermentas, Lithuania). PCR was performed using the following cycling parameters: a cycle denaturation (94°C, 2 min); 30 cycles of denaturation (94°C, 30 s), annealing (55°C, 1 min) and extension (72°C, 90 s), followed by a final extension step (72°C, 10 min). The PCR products were analyzed on 1.5% (w/v) agarose gel in Tris-acetate-EDTA buffer. The gels were stained with 0.2 µg/mL of ethidium bromide (Amresco Cat no. 0492, USA) and photographed under UV light. Sequencing of the PCR products was done by Ref Gen Ltd. (Ankara University Technopolis, Turkey) using an automated gene sequencer ABI PRISM 3730XL (Perkin Elmer, USA). The sequences of the PCR products were compared to the 16S rDNA gene sequences of Genbank using the BLAST program for detection of the closest relatives. Phylogenetic analysis was conducted with MEGA software version 4.0 (MEGA; http://www.megasoftware.net). The tree was generated by neighbor-joining using the maximum composite likelihood model (Tamura et al., 2007).
Antibiotic resistance patterns of isolates were detected by the disc diffusion method on Mueller-Hinton agar (Cariolato et al., 2008). A total of 18 commercial antibiotic discs of ampicillin (10 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), erythromycin (15 μg), gentamicin (120 μg), levofloxacin (5 μg), linezolid (30 μg), minocycline (30 μg), nitrofurantoin (300 μg), norfloxacin (10 μg), penicillin G (10 U), quinupristin/dalfopristin (15 μg), rifampin (5 μg), streptomycin (300 μg), teicoplanin (30 μg), tetracycline (30 μg) and vancomycin (30 μg) were used. All antibiotics were obtained from Oxoid Ltd. (UK). Susceptibility or resistance of enterococci was determined according to the Clinical and Laboratory Standards Institute (2012).
The presence of chloramphenicol (cat), ciprofloxacin (gyrA), erythromycin (ermA, ermB, ermC), tetracycline (tetM, tetL, tetK, tetS, tetO) and vancomycin (vanA, vanB, vanC) resistance genes in enterococci was investigated by PCR. PCR primers and annealing temperatures for detection of antibiotic resistance genes are listed in Table 1. PCR was performed in 50 μL reaction mixtures. PCR conditions involved the following cycling parameters: initial denaturation cycle at 94°C for 2 min (95°C for 5 min for gene cat), next 30 cycles of denaturation at 94°C for 60 s (95°C for 30 s for gene cat), annealing at an appropriate temperature for 60 s (30 s for gene cat) and elongation at 72°C for 60 s (30 s for gene cat), and a final extension cycle at 72°C for 10 min (7 min for gene cat) (Dutka-Malen et al., 1995; Kim et al., 2005; Ouoba et al., 2008; Reviriego et al., 2005).
| Genes | Primer sequence (5' to 3') | Annealing temperature (°C) | Reference |
|---|---|---|---|
| cat | GCGAACGAAAAACAATTGCA | 55 | (Kim et al., 2005) |
| TGAAGCTGTAAGGCAACTGG | |||
| gyrA | GAYTATGCWATGTCAGTTATTGT | 45 | (Ouba et al., 2008) |
| GGAATRTTRGAYGTCATACCAAC | |||
| ermA | AAGCGGTAAAACCCCTCTGAG | 55 | (Ouba et al., 2008) |
| TCAAAGCCTGTCGGAATTGG | |||
| ermB | CATTTAACGACGAAACTGGC | 52 | (Ouba et al., 2008) |
| GGAACATCTGTGGTATGGCG | |||
| ermC | CAAACCCGTATTCCACGATT | 48 | (Ouba et al., 2008) |
| ATCTTTGAAATCGGCTCAGG | |||
| tetM | GTTAAATAGTGTTCTTGGAG | 45 | (Ouba et al., 2008) |
| CTAAGATATGGCTCTAACAA | |||
| tetL | GTTGCGCGCTATATTCCAAA | 54 | (Ouba et al., 2008) |
| TTAAGCAAACTCATTCCAGC | |||
| tetS | TGGAACGCCAGAGAGGTATT | 55 | (Ouba et al., 2008) |
| ACATAGACAAGCCGTTGACC | |||
| tetK | TTAGGTGAAGGGTTAGGTCC | 55 | (Ouba et al., 2008) |
| GCAAACTCATTCCAGAAGCA | |||
| tetO | GATGGCATACAGGCACAGAC | 55 | (Ouba et al., 2008) |
| CAATATCACCAGAGCAGGCT | |||
| vanA | GGGAAAACGACAATTGC | 54 | (Dutka-Malen et al., 1995) |
| GTACAATGCGGCCGTTA | |||
| vanB | GTGCTGCGAGATACCACAGA | 54 | (Reviriego et al., 2005) |
| CGAACACCATGCAACATTTC | |||
| vanC | GGTATCAAGGAAACCTC | 54 | (Dutka-Malen et al., 1995) |
| CTTCCGCCATCATAGCT |
Y, C or T; R, A or G; W, A or T.
Results and Discussion
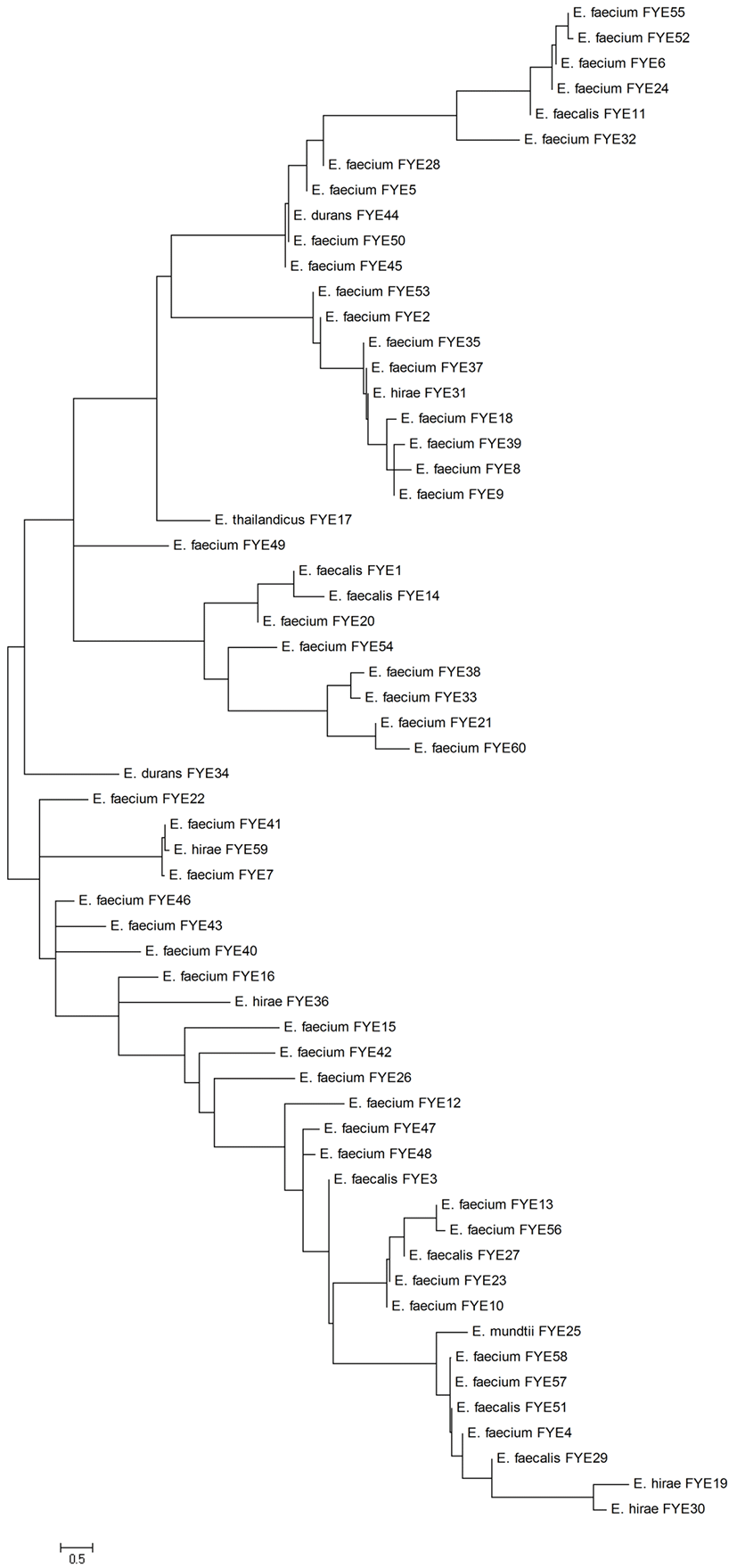
A total of 63 presumptive enterococci isolates, which appear surrounded by a black halo on KAA agar, were isolated from 20 Sucuk samples obtained from Afyonkarahisar province, Turkey. Three isolates Gram-positive rods were eliminated and the remaining 60 isolates Gram-positive cocci (single, pairs or in short chains) were selected for further analyses. All the Gram-positive cocci isolates were catalase-negative and resistant to heat at 60°C for 30 min. In addition, they were grown in MRS broth at 10°C, 45°C, pH 9.6 and presence of 6.5% NaCl. The results of Gram staining, catalase and cultural tests showed that these 60 strains isolated from Sucuk samples were members of the Enterococcus genus. 60 presumptive Enterococcus strains were identified at species level by 16S rDNA sequence analyses. Presumptive Enterococcus strains were identified as 44 E. faecium (73.33%), 7 E. faecalis (11.67%), 5 E. hirae (8.33%), 2 E. durans (3.33%), 1 E. mundtii (1.67%) and 1 E. thailandicus (1.67%). Phylogenetic analysis of 16S rRNA gene sequences of Enterococcus strains was shown in Fig. 1. Enterococci are associated with some traditional sausages produced in different European countries (Jahan et al., 2013; Landeta et al., 2013; Yüceer and Özden Tuncer, 2015), as confirmed in this study. Dominant Enterococcus species in Sucuk samples were determined as E. faecium (73.33%). Similar to our results, most previous studies reported E. faceium as a dominant microbiota in fermented sausages (Landeta et al., 2013; Valenzuela et al., 2009; Yüceer and Özden Tuncer, 2015). In contrast, some researchers found a higher percentage of E. faecalis than other species in animal originated foods (Jahan et al., 2013; Peters et al., 2003).
Sixty Enterococcus strains were tested for their resistance to 18 different antibiotics using the disc diffusion method. Antibiotic resistance test results of Enterococcus strains were given in Table 2. Only E. faecium FYE4 and FYE60 strains displayed susceptibility to all antibiotics. All of the strains were found sensitive only to ampicillin. Similar to our result, some researchers showed that enterococci isolated from different traditional fermented sausages and cheeses were completely sensitive to ampicillin (Jahan et al., 2013; Landeta et al., 2013; Yogurtcu and Tuncer, 2013; Yüceer and Özden Tuncer, 2015). On the other hand, Chajecka-Wierzchowska et al. (2012) showed that low numbers of enterococci isolated from animal originated food were resistant to ampicillin. Enterococcus strains showed different susceptibility patterns to the other antibiotics used in this study. Antibiotic susceptibility and resistance percentage of Enterococcus strains were given in Table 3.
1)AMP, Ampicillin (10 μg); C, Chloramphenicol (30 μg); CIP, Ciprofloxacin (5 μg); CN, Gentamisin (120 μg); DO, Doxycycline (30 μg); E, Erythromycin (15 μg); F, Nitrofurantoin (300 μg); LEV, Levofloxacin (5 μg); LZD, Linezolid (30 μg); MH, Minocycline (30 μg); NOR, Norfloxacin (10 μg); P, Penicillin G (10 U); QD, Quinupristin/dalfopristin (15 μg); RD, Rifampin (5 μg); S, Streptomycin (300 μg); TEC, Teicoplanin (30 μg); TE, Tetracycline (30 μg); VA, Vancomycin (30 μg).
2)S, susceptible; I, intermediary; R, resistance.
1)Penicillin G 10 U/disc.
Most of the strains were found sensitive to gentamicin (93.33%), streptomycin (93.33%), chloramphenicol (90.00%), minocycline (90.00%), penicillin G (90.00%), doxycycline (88.33%), tetracycline (88.33%) and vancomycin (86.67%), followed by other antibiotics. On the contrary, Enterococcus strains were found resistant to rifampin (51.67%), ciprofloxacin (38.33%), nitrofurantoin (33.33%) and erythromycin (21.67%), followed by other antibiotics except for ampicillin. Recently, similar observations to our results have been reported by several researchers. Peters et al. (2003) indicated that 22.03% of the Enterococcus strains isolated from animal originated foods were sensitive to erythromycin. Chajecka-Wierzchowska et al. (2012) reported that enterococci from food of animal origin were sensitive to chloramphenicol (96.73%), streptomycin (91.30%), gentamicin (85.86%) and quinupristin/dalfopristin (66.66%). In addition, Yüceer and Özden Tuncer (2015) showed that Enterococcus strains from Sucuk were sensitive to chloramphenicol (100%), doxycycline (100%), gentamicin (100%), minocycline (100%), streptomycin (100%), tetracycline (100%), penicillin G (88%), levofloxacin (52%), rifampin (28%) and ciprofloxacin (12%). Conversely to our results, some researchers showed that Enterococcus strains isolated from animal originated foods were 100% sensitive to levofloxacin, quinupristin/dalfopristin, teicoplanin or vancomycin (Chajecka-Wierzchowska et al., 2012; Landeta et al., 2013; Peters et al., 2003; Yogurtcu and Tuncer, 2013; Yüceer and Özden Tuncer, 2015).
Antibiotic susceptibility and resistance percentage of enterococci at species level was shown in Table 4. Among the Enterococcus species, E. faecalis showed the highest percentage of resistance to the tested antibiotics. E. faecalis strains displayed resistance to quinupristin/dalfopristin (85.71%), erythromycin (71.43%), chloramphenicol (57.14%) and streptomycin (57.14%). On the other hand, E. faecium strains were found resistant to rifampin (56.82%), ciprofloxacin (45.45%) and nitrofurantoin (34.09%). E. hirae strains showed resistance only to nitrofurantoin. Three out of five E. hirae strains were found resistant to nitrofurantoin. Both of the E. durans strains isolated from Sucuk exhibited resistance to rifampin. In addition, E. durans FYE44 was found resistant to erythromycin, nitrofurantoin, penicillin G and teicoplanin. E. mundtii FYE25 did not display resistance to any antibiotics used in this study. E. thailandicus FYE17 showed resistance to erythromycin and rifampin. Similar to our results, Yogurtcu and Tuncer (2013) reported that E. faecalis strains isolated from Turkish tulum cheese were found more resistant to antibiotics than other species. On the other hand, Chajecka-Wierzchowska et al. (2012) determined that E. faecalis and E. faecium strains isolated from ready-to-eat food of animal origin showed similar antibiotic resistance patterns. Conversely, some researchers reported that E. faecium strains were more resistant to antibiotics than other species (Landeta et al., 2013; Valenzuela et al., 2009).
1)n, number of strain.
2)S, susceptible; I, intermediary; R, resistance.
The Enterococcus strains (61.67%) displayed resistance from between two and eight antibiotics. Some of the strains (11.67%) showed resistance to ciprofloxacin and rifampin. Multiple antibiotic resistance in enterococci is not surprising. Several researchers reported that multiple antibiotic resistance in enterococci isolated from fermented meat products is common (Jahan et al., 2013; Yüceer and Özden Tuncer, 2015), as confirmed in this study. Jahan et al. (2013) showed that Enterococcus strains isolated from fermented meat products exhibited a high rate (93.10%) of multiple antibiotic resistance characteristics. Researchers also reported that 58.62% of the strains displayed resistance from between three and eight antibiotics. In addition, Yüceer and Özden Tuncer (2015) determined that 68% of the Enterococcus strains isolated from fermented Sucuk exhibited multiple antibiotic resistance. The strains displayed resistance from between two and five antibiotics.
A total of 60 Enterococcus strains were evaluated for the presence of chloramphenicol (cat), ciprofloxacin (gyrA), erythromycin (ermA, ermB, ermC), tetracycline (tetM, tetL, tetK, tetS, tetO) and vancomycin (vanA, vanB, vanC) resistance genes. Thirty-one out of 60 isolates showed positive PCR for gyrA, ermB, ermC, tetM, tetL and vanA. A lack of correlation between genotypic and phenotypic analysis was detected (Table 2).
A correlation between the chloramphenicol resistance phenotype and genotype was not detected in the researched strains. The cat gene was not detected by PCR in five phenotypically chloramphenicol-resistant strains. This result indicated that other chloramphenicol resistance genes may be influenced in these strains. From this, we presumed that positive PCR results for cat gene were not detected in these strains by the primers used in this study. Similar to our results, Kim et al. (2005) determined that 17 out of 29 chloramphenicol-resistant Staphylococcus intermedius isolates contained the cat gene, while 12 isolates did not carry the cat gene by the same primer set used in our study. In addition, Jahan et al. (2013) reported that only E. faecium S15 strain was positive for the cat gene among three phenotypically chloramphenicol-resistant enterococci isolated from meat and fermented meat products.
The gyrA gene was detected in nine Enterococcus strains that are phenotypically one resistant, seven intermediate and one susceptible). Only one strain, E. faecium FYE5, was given a positive result for gyrA gene among 23 phenotypically ciprofloxacin-resistant Enterococcus strains. Similar to our results, Jahan et al. (2013) did not detect gyrA or parC gene in any of three ciprofloxacin-resistant Enterococcus strains isolated from meat and fermented meat products. In addition, PCR results showed that phenotypically ciprofloxacin-susceptible E. hirae FYE36 strain contains the gyrA gene (Table 2). In Gram positive bacteria, both gyrA and parC genes are defined to be the primary target in quinolone resistance. Mutations in Ser84 locus of GyrA in Staphylococcus aureus resulted in increased of ciprofloxacin-resistance. Similarly, single mutations taking place in the Ser83 and the Glu87 loci of GyrA were related to an increase of minimal inhibitory concentration values of ciprofloxacin in E. faecalis strains. The mutations taking place in both gyrA and parC genes were found in high-level quinolone-resistant E. faecium (Petersen et al., 2004).
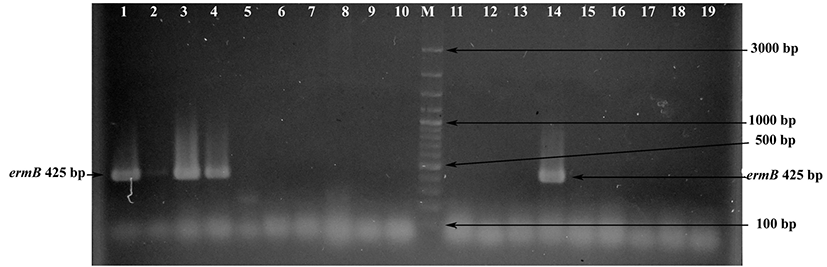
The erythromycin resistance gene ermA was not detected in any strains. However, ermB and ermC genes were detected in seven (11.67%) and 13 (21.67%) strains, respectively. These strains were showed the expected amplification fragments of 425 bp (Fig. 2) and 295 bp when their DNAs were amplified with specific primers for ermB and ermC genes, respectively. Conversely to our results, previous studies showed that the ermB gene is the most common erythromycin-resistant gene defined in Enterococcus species (Aarestrup et al., 2000; Jahan et al., 2013). In our study, the ermC gene was found at a higher frequency than the ermB gene. A lack of correlation between phenotypic and genotypic analysis for erythromycin resistance was detected (Table 2). The ermB gene was detected in six erythromycin-resistant and one erythromycinsusceptible Enterococcus strains. In addition, the ermC gene was found in nine erythromycin-intermediate, two erythromycin-resistant and one erythromycin-susceptible strains. The erythromycin-resistant strains E. faecium FYE7, E. thailandicus FYE17, E. durans FYE44 and E. faecium FYE45 did not present any of the three erm genes (Table 2), indicating that other factors influence resistance in these strains. Similar to these results, Jahan et al. (2013) demonstrated the presence of ermB and/or mefA/B genes in seven of 13 phenotypically erythromycin-resistant Enterococcus strains isolated from meat and fermented meat products. However, erm genes (ermA, ermB, ermC or mefA/B) were not found in the remaining six Enterococcus strains. In our study, it is interesting that the presence of ermB and ermC genes was determined in phenotypically erythromycin-susceptible E. faecium FYE4 and E. faecium FYE16 strains, respectively. A similar observation was recently reported by Ding et al. (2012). In this study, researchers determined the presence of ermB and ermB/ermC genes in six and two erythromycin-susceptible Staphylococcus strains, respectively. The researchers showed that the mutation of 23S rRNA gives rise to many erythromycin-susceptible isolates when it has erm genes, the mutation on 23S rRNA possibly disturbing the site of the methylation, allowing erythromycin to bind to the ribosome. The reason for lack of correlation between genotypic and phenotypic analysis in E. faecium FYE4 and E. faecium FYE16 strains may be due to mutation of the 23S rRNA gene in these strains. Further investigations are needed for the determination of erythromycin susceptibility mechanisms in these strains.
The tetracycline resistance genes tetM and tetL were detected in seven (11.67%) and four (6.67%) strains, respectively. In tetracycline-resistant E. faecium FYE32 and E. faecium FYE37 strains carried the tetL gene and E. faecalis FYE14 strain carried both tetM and tetL genes. Moreover, tetM gene was detected in three out of four tetracycline-intermediate strains. The tetK, tetS and tetO genes were not detected in any strains. Different researchers have reported that the tetM gene is more common in tetracyclineresistant Enterococcus strains (Aarestrup et al., 2000; Cauwerts et al., 2007; Jahan et al., 2013), as confirmed in this study. Similar to our results, Aarestrup et al. (2000) detected tetM and tetL genes in tetracycline-resistant E. faecalis and E. faecium strains isolated from humans, broilers and pigs. Researchers did not detect tetK and tetS genes in any animal-origin isolates. Hummel et al. (2008) reported that 94%, 63% and 56% of tetracycline-resistant Enterococcus strains isolated from cheese contained tetL, tetM and tetK genes, respectively. In addition, researchers reported that none of the strains contained tetO or tetS genes, as confirmed in this study. In another study, Jahan et al. (2013) also detected tetM (16 strains), tetL (six strains), and tetK (six strains), genes in 19 Enterococcus strains isolated from meat and fermented meat products, respectively. The tetO and tetS genes were not found in any of the strains. A lack of correlation between phenotypic and genotypic analysis for tetracycline resistance was detected (Table 2). In our study, the presence of both tetM and tetL genes was determined in the phenotypically tetracycline-susceptible E. faecium FYE41 strain. Similar observations were detected by Cauwerts et al. (2007). Researchers determined that two strains contained tetM and one strain contained tetL among three phenotypically tetracycline-sensitive E. faecium strains isolated from broilers.
The vancomycin resistance genes vanB and vanC were not detected in any tested strains. However, vanA gene was detected only in vancomycin-intermediate E. faecium FYE16 strain. One vancomycin-resistant and six vancomycin-intermediate Enterococcus strains did not present any of the three vancomycin resistance genes (Table 2). The vanA is the most common glycopeptide resistance gene in enterococci and it is usually related to high-level vancomycin resistance. Most of the VanA type resistant strains are also teicoplanin resistant (Garrido et al., 2014). In this study, the phenotypically vancomycin-resistant strain, E. faecalis FYE29, was also found to be teicoplanin-resistant but, none of the vancomycin resistance genes researched in this study were found, indicating that other factors influence resistance in this strain. Baylan et al. (2011) reported that one out of eight phenotypically vancomycin- and teicoplanin-resistant urinary E. faecium isolates did not contain vanA or vanB genes, as confirmed in this study. The vanA, vanB and vanC genes were detected in none of the phenotypically vancomycin-sensitive Enterococcus strains as expected. Conversely to our findings, Szakacs et al. (2014) determined the presence of the vanA gene in clinical isolate of vancomycin-sensitive E. faecium.
Conclusion
The results of this study indicated that E. faecium is the dominant Enterococcus species present in Sucuk. Enterococcus strains were found resistant to the clinically relevant antibiotics except ampicillin. Most of Enterococcus strains displayed multiple antibiotic resistance. The ermB, ermC, gyrA, tetM, tetL and vanA genes were detected in some strains. These strains may play a role in the spread of antibiotic resistance among bacteria, and so could create a health risk for consumers. The results of this study indicated that Sucuk manufactured without using a starter culture is a reservoir of multiple antibiotic resistant enterococci. Consequently, Sucuk is a potential reservoir for the transmission of antibiotic resistance genes from animals to humans. Other potential risk factors (biogenic amine production and virulence factors) of enterococci isolated from Sucuk should be researched to protect consumer health. Moreover, further investigations are also needed for the determination of the lack of correlation between antibiotic resistance genotypes and phenotypes in enterococci to clarify antibiotic resistance and susceptibility mechanisms of these bacteria.













