Introduction
Escherichia coli are facultative anaerobic bacteria found in human intestines, and can be categorized into pathogenic and non-pathogenic E. coli. The clinical symptoms of pathogenic E. coli infection are diarrheal disease, urinary tract infection, sepsis, and meningitis (Nataro and Kaper, 1998). Pathogenic E. coli are categorized as enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), and enterohemorrhagic E. coli (EHEC), according to their pathogenic features (Kuhnert et al., 2000; Sidhu et al., 2013). In South Korea, there were 38 E. coli outbreaks in 2006 and 59 E. coli outbreaks in 2016 (MFDS, 2017). During 2004-2006, Lee et al. (2009) isolated EHEC, ETEC, and EPEC strains from pork (201/3000; 14.9%), beef (31/3000; 4.1%), and poultry (41/3000; 4.6%), purchased from meat processing facilities, butcher shops, wholesale stores, and wet markets in several regions of South Korea. Recently, E. coli outbreaks resulting from vegetable consumptions have also been reported (CDC, 2017; Denis et al., 2016; Hong et al., 2012; Lee et al., 2016; Wendel et al., 2009). Consumption of baby leafy vegetables has increased, usually in the form of fresh-cut salad, which is vulnerable to contamination with pathogens from soil, water, and human handling (Beuchat et al., 1996; Tomás-Callejas et al., 2011).
The traditional method for isolation and identification of pathogens involves a lot of time and labor (Fukushima et al., 2007). Rapid identification of pathogens is important to prevent foodborne illnesses, and a rapid detection method is therefore necessary, especially for raw foods (Ge and Meng, 2009; Naravaneni and Jamil, 2005). A rapid detection method should be sufficiently sensitive to detect low concentrations of pathogens, and should be time-efficient for immediate detection (Law et al., 2015). However, rapid detection methods are not usually accurate at low cell concentrations (Mandal et al., 2011). If the bacterial cells on foods can be concentrated by a physical method, rather than by enrichment, the speed of rapid detection methods may be improved (Stevens and Jaykus, 2004). Polymerase chain reaction (PCR) is a commonly used nucleic acid-based method, which has high sensitivity and specificity compared to that of immunological methods based on antibody-antigen reactions (Law et al., 2015).
Therefore, the objective of this study was to evaluate the efficiency of centrifugation and filtration pretreatment methods to decrease sample preparation time and to improve the sensitivity and specificity of multiplex PCR for the rapid detection of low concentrations of pathogenic E. coli in various foods.
Material and Methods
A single colony of each pathogenic E. coli strain [E. coli NCCP11142, E. coli NCCP14037, E. coli NCCP 14038, E. coli NCCP14039, and E. coli NCCP15661] was cultured in 10 mL tryptic soy broth (TSB; Becton, Dickinson and Company, USA) at 37°C for 24 h. A 0.1 mL aliquot was then subcultured in 10 mL TSB at 37°C for 24 h. A five-strain mixture (25 mL) of pathogenic E. coli was centrifuged at 1,912 g and 4°C for 15 min, and the cell pellet was washed twice with phosphate-buffered saline (PBS, pH 7.4; 0.2 g of KH2PO4, 1.5 g of Na2HPO4·7H2O, 8.0 g of NaCl, and 0.2 g of KCl in 1 L of distilled water) and then diluted serially to obtain 3, 4, and 5 Log CFU/mL.
Samples (pork, beef, and baby leafy vegetables) were purchased from a butcher shop and a market located in Seoul, South Korea in March, 2017. Pork (leg butt), beef (leg butt), and baby leafy vegetables were purchased and tested for the presence of E. coli. Twenty five grams of pork, 25 g of beef and 10 g of baby leafy vegetables were placed into a filter bag (3M, USA), and 0.1 mL of bacterial suspension was inoculated onto each sample to obtain 1, 2, and 3 Log CFU/g. A total of four samples (n=4) were prepared for each inoculum concentration. After inoculation, the samples were massaged 20 times to allow E. coli attachment.
To enumerate E. coli cells in the inoculated samples, negative control samples and E. coli inoculated samples at 1, 2, and 3 Log CFU/g were resuspended with 50 mL 0.1% BPW in a filter bag, and shaken 30 times. One-milliliter aliquots of the diluents were then spread onto E. coli/Coliform Count Plate (Petrifilm™, 3M, USA). The plates were incubated at 37°C for 24 h and blue colonies with gas bubbles were then counted. Four replicates (n=4) were analyzed for bacterial concentrations and sample pretreatments.
For each sample in bags as previously described, bacteria were allowed to be attached for 15 min, and then 50 mL of 0.1% buffered peptone water (BPW; Becton, Dickinson and Company) was added and shaken 30 times. The shaken samples were pretreated as follows: (i) control: shaken sample, (ii) centrifuged: 50 mL of the shaken sample was transferred to a conical tube and centrifuged at 1,912 g at 4°C for 15 min, and the cell pellet was then resuspended in 5 mL 0.1% BPW, and (iii) filtered: 50 mL of the shaken sample was filtered through filter paper (Hyundai micro Co., Ltd., Korea) and subsequently through a 0.45 µm membrane filter (Hyundai micro Co., Ltd.), followed by vortexing the membrane filter in 5 mL 0.1% BPW for 1 min.
One-milliliter aliquots of control, centrifuged, and filtered samples were used for DNA extraction. DNA was extracted with PowerprepTM DNA Extraction Food and Feed Kit (KogeneBiotech, Korea) according to the manufacturer’s protocol, and the DNA extracts were subjected to PCR with PowerchekTM Diarrheal E. coli 8-plex Detection Kit (KogeneBiotech) according to the manufacturer’s protocol. Eight genes [heat-labile enterotoxin (LT; 530 bp), heat-stable enterotoxin (ST; 167 bp), bfpA (400 bp), eaeA (231 bp), aggR (757 bp), stx1 (637 bp), stx2 (297 bp), and ipaH (141 bp)] can be detected by this kit.
Results and Discussion
The absence of E. coli in the initial pork, beef, and baby leafy vegetable samples was confirmed by a quantitative experiment (detection limit: 0.5 Log CFU/g for pork and beef, and 0.8 Log CFU/g for baby leafy vegetables). To evaluate the effect of centrifugation and filtration on decreasing sample preparation time and improving the sensitivity of PCR, pathogenic E. coli were inoculated into the samples at 1, 2, and 3 Log CFU/g, and targeted E. coli concentrations were obtained (Table 1). The presence of pathogenic E. coli was detected by PowerchekTM Diarrheal E. coli 8-plex Detection Kit. The kit can detect eight genes, including heat-labile enterotoxin gene (LT; 530 bp), heat-stable enterotoxin gene (ST; 167 bp), bfpA (400 bp), eaeA (231 bp), aggR (757 bp), stx1 (637 bp), stx2 (297 bp), and ipaH (141 bp). E. coli genes were detected in the inoculum using the kit. Five of the tested E. coli strains possessed the genes eaeA (E. coli NCCP 11142, 14037, 14038, and 15661), aggR (E. coli NCCP 14039), and bfpA (E. coli NCCP 15661).
1),2): detection limit
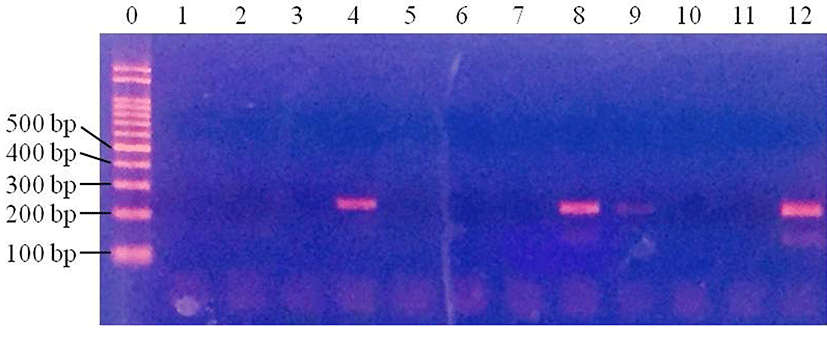
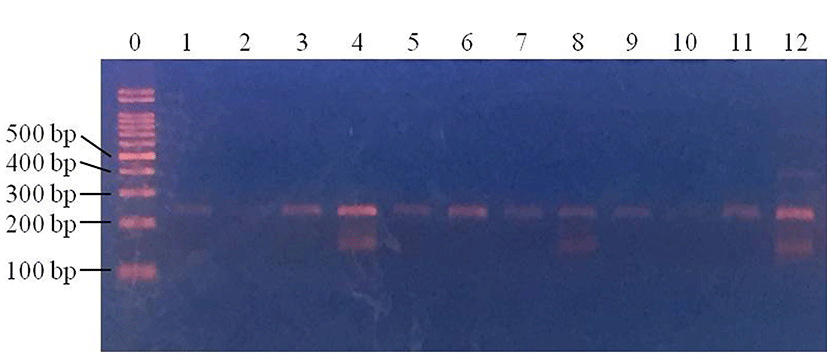
After pretreatment of inoculated samples, PCR was performed to compare the effect of sample pretreatment on the time taken to detect pathogenic E. coli. In pork samples, pathogenic E. coli were not detected in the control samples at any of the inoculum concentrations (data not shown). Centrifuged pork samples were all E. coli-positive (100%; 4/4) at 3 Log CFU/g, while no E. coli-positive samples were observed at 1 and 2 Log CFU/g (p<0.05) (Fig. 1). For filtered pork samples, E. coli was detected in all samples (100%; 4/4) at 3 Log CFU/g and 2 Log CFU/g (100%; 4/4), but at 1 Log CFU/g, no E. coli was detected in any of the samples (p<0.05) (Fig. 2). This result indicates that filtration pretreatment can decrease sample preparation time, compared to enrichment because filtration takes less than 30 min, but enrichment takes 24 h as well as improving the sensitivity of PCR when detecting E. coli in pork.
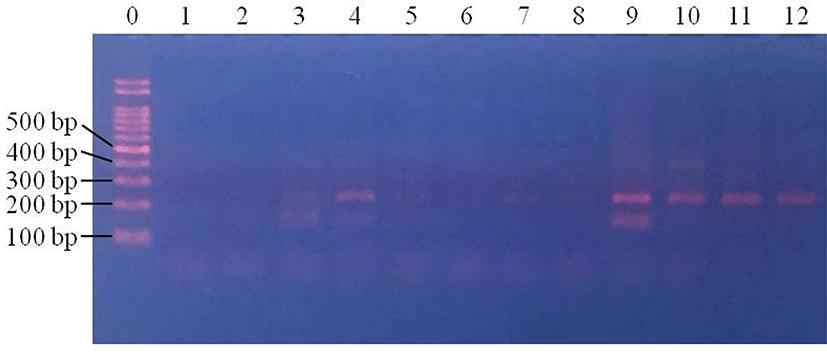
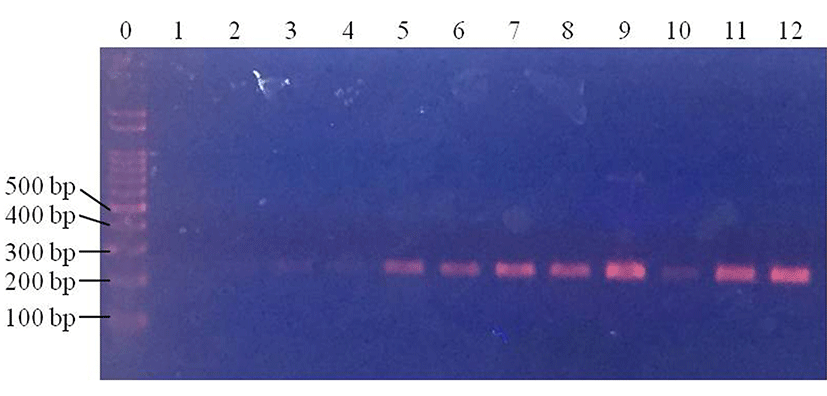
In beef samples, E. coli was not detected in control samples (data not shown). Of the four centrifuged samples, E. coli was detected in three (75%; 3/4) samples at 1 Log CFU/g (Fig. 3). E. coli was detected in two of the centrifuged samples at 2 Log CFU/g (50%; 2/4), and in three samples at 3 Log CFU/g (75%; 3/4) (Fig. 3). In the filtered samples, E. coli was detected at 2 and 3 Log CFU/g, but not at 1 Log CFU/g (p<0.05) (Fig. 4). This result suggests that even though centrifugation can allow the detection of E. coli at 1 Log CFU/g in beef, the detection rates were not 100% at any concentrations. On the other hand, filtration allowed the detection of 2 and 3 Log CFU/g E. coli in 100% of the samples. Thus, filtration pretreatment can support rapid pathogenic E. coli detection in beef, using PCR.
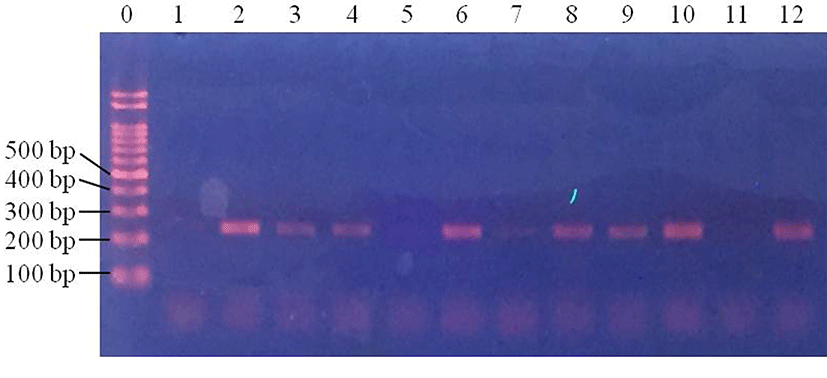
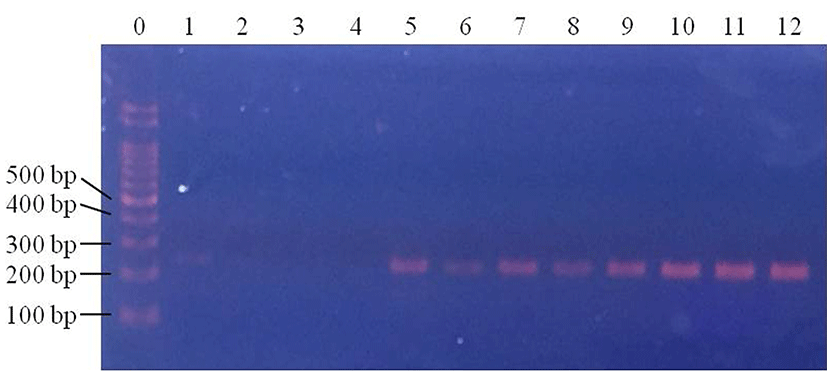
In baby leafy vegetables, E. coli were detected in the control samples in 25% of 1 Log CFU/g samples, 25% of 2 Log CFU/g samples and 50% of 3 Log CFU/g samples (Fig. 5). In samples subjected to centrifugation, E. coli was detected in 0% of 1 Log CFU/g samples, 25% of 2 Log CFU/g samples and 25% of 3 Log CFU/g samples (Fig. 6). Following filtration, E. coli was detected in 75% of 1 Log CFU/g, 100% of 2 Log CFU/g, and 100% of 3 Log CFU/g samples (Fig. 7). This indicates that filtration is a suitable pretreatment, which contributes to rapid detection of pathogenic E. coli by PCR.

Enrichment, although time-consuming, is necessary prior to detection of pathogens by PCR, especially for low concentrations of bacteria (Fukushima et al., 2007; Stevens and Jaykus, 2004). During enrichment, other bacteria are also enriched, especially in meat that contains more endogenous bacteria than vegetables, and the characteristics of the food matrix could affect the PCR assay (Bhunia, 2014; Wang and Salazar, 2016). PCR has high sensitivity and high specificity for the detection of foodborne pathogens (Law et al., 2015), but in combination with enrichment, it cannot be a rapid detection method. Therefore, pretreatment for rapid detection using PCR is critical as described, and according to the results from our study, it can be concluded that filtration pretreatment can replace enrichment prior to PCR for the detection of pathogenic E. coli in pork, beef, and baby leafy vegetables, and can thus save time when detecting even low concentrations of E. coli.













