Introduction
In China, lamb meat is favored by most of the consumers due to its delicious taste and low cholesterol content. Further, the yield of lamb has reached 4.92 million tons with an increasing trend (Meng and Zhang, 2018). Lamb meat with high nutrient and water contents is susceptible to microbial spoilage, leading to meat quality decline, food safety issues, and economic loss (Gram et al., 2002).
The growth of microorganisms and their metabolic activity can significantly affect the shelf life of lamb meat. Analysis of microbial counts and community changes during the storage may contribute to the inhibition of specific spoilage and the extension of shelf life. Several investigations of microbial communities were performed on lamb meat in Iran (Sani et al., 2017), USA (Kim et al., 2015), Australia (Kiermeier et al., 2013), Spain (Oses et al., 2013), and China (Wang et al., 2016; Zhou et al., 2015). Although different package methods or storage conditions were involved in different studies, the dominating spoilage bacteria of refrigerated lamb meat were identified as lactic acid bacteria (LAB), Brochothrix, Enterobacteriaceae, and Pseudomonas (Mills et al., 2014; Ortuno et al., 2017; Oses et al., 2013). Furthermore, lamb can be contaminated with foodborne pathogens such as Shiga toxin-producing Escherichia coli, indicating that the microbial detection of pathogens is important for assuring meat safety (Oses et al., 2013). Due to the higher pH (≥5.8) and nutritional composition of lamb meat (Mills et al., 2014), variations in bacterial communities were observed in lamb compared with other meat products, such as pork and beef. For example, certain species of Enterobacteriaceae were able to grow aerobically/ anaerobically on adipose and muscle tissues with high pH (> 6), and were prevalent on lamb (Sun and Holley, 2012).
Previous studies reported varied microbial communities of lamb meat. The feeding, slaughter environment and practices, cross-contamination initialized bacteria loading of lamb meat, meanwhile water activity, meat pH, storage temperature, and packaging form affect the dynamics of microflora in lamb meat (Mills et al., 2014; Ortuno et al., 2017). These factors consequently impact the shelf life of raw meat. Packaging is the most commonly used method of preserving meat that prolongs the shelf life. Like other red meat, air pack (plastic tray wrapped with polyethylene), vacuum pack, and modified atmosphere pack are most commonly used package types in chilled lamb. Compared with air pack, vacuum and modified atmosphere packaging can significantly delay the growth rate of microorganisms in chilled lamb, especially aerobic microorganisms, such as Pseudomonas and Acinetobacter, thus significantly prolonging the shelf life of lamb (Newton et al., 1977; Soldatou et al., 2009). Our previous study showed that high CO2 atmosphere (100%) inhibited the microbial growth of LAB, Pseudomonas spp, and Enterobacteriaceae and also delayed changes in the microbial community composition, hence extending the lamb’s shelf life by approximately 7 days compared with the vacuum package (Wang et al., 2016).
Air pack in a plastic tray (PT) is the predominant packing form of chilled lamb in China. However, a limited number of studies focused on bacterial communities of lamb in this packaging form during chill storage, except Zhou et al. (2015) who reported that the main bacteria were Pseudomonas, Bacillus, and Acinetobacter using colony polymerase chain reaction (PCR). Insufficient investigation on global bacterial communities of air pack may mislead the preserving method for chill lamb in China. In this study, Polymerase chain reaction–denaturing gradient gel electrophoresis (PCR-DGGE) was employed to investigate the dynamics of bacterial communities of the chilled lamb packaged in a PT and a vacuum pouch (VAC). during the storage period. The data could furthermore provide an insight to improve the lamb hygiene and prolong the shelf life of the lamb.
Materials and Methods
The hind legs of 10 fresh lambs, provided by the local abattoir (Beijing Zhuochen, China), were cut into 42 steaks and transported to the laboratory on ice in a foam box within 2 h. Twenty-one lamb steaks were individually put into a sterilized PT covered with a conventional polyethylene wrap film surrounded by the air atmosphere. Twenty-one samples were individually packaged in a VAC with a vacuum-packaging machine (Jiaxing Expro Stainless Steel Engineering Co., Ltd, China). All of the samples were stored under chilled conditions where the temperature ranged from 4°C to 6°C. The lamb steaks stored in a PT were subjected to analysis on Days 0, 1, 2, 3, 4, 5, 6, and 7 over the total storage period. For the lamb packaged in a VAC, the lamb steaks were analyzed on Days 0, 3, 6, 9, 12, 15, 18, and 21. The analysis of the samples on Day 0 prior to packaging was the same for both the two groups.
Twenty-five grams of lamb meat from each sample was diluted in 225 mL of sterile saline (8.5 g/L NaCl) and then homogenized for 1 min with a stomacher aseptically. The serial decimal dilutions were prepared, and suitable dilutions were incubated on the plate count agar (PCA, Aoboxing, Beijing, China) at 37°C for 48 h in duplicate to determine the counts of viable bacteria. After incubation, the numbers of colony-forming units (CFU) per gram of meat were calculated as total viable counts (TVC).
Ten grams of lamb meat cut from different parts of the sample were put into a sterile tube with 20 mL of phosphate-buffered saline (pH 7.4) solution and agitated for 5 min. Then, the tube was centrifuged for 5 min at 200 g at 4°C to separate the large debris. Next, the supernatant was removed to a new sterile tube for further centrifugation at 12,000 g at 4°C for 10 min to harvest the bacterial precipitate applied for extracting DNA (Jiang et al., 2010). Bacterial genomic DNA was extracted using the phenol–chloroform method as described previously (Via and Falkinham, 1995).
The primers 341F-GC (5′-CGCCCGCCGCGCGCGGCGGGCGGG GCGGGGGCACGGGGGGCCTACGGGAGGCAG CAG-3′) and 534R (5′-ATTACCGCGGCTGCTGG-3′) were used to amplify V3 variable region of 16S rDNA gene (Muyzer et al., 1993), following the program as described by Lubbs et al. (2009) with a PTC-200 Peltier Thermo Cycler (Bio-Rad, CA, USA). The products of the amplification with the length of approximately 200 bp were inspected by electrophoresis in 2% (w/v) agarose gels. Then, DGGE was performed using a DCode Universal Mutation Detection System (Bio-Rad). The PCR products were separated on 8% (w/v) polyacrylamide gel using a 30%–60% linear denaturing gradient. The gel was subjected to a constant voltage of 150 V for 10 h at 60°C in the 0.5× TAE solution (20 mM Tris-acetate, pH 7.4, 10 mM sodium acetate, 0.5 mM Na2-EDTA) (Muyzer et al., 1993). Gels were stained with GelGreen (Biotium, CA, USA) and imaged using an Amersham Imager 600 (GE Healthcare, IL, USA).
The selected bands were punched with a sterile scalpel and the DNA was extracted using a Poly-Gel DNA Extraction Kit (Solarbio, Beijing, China) to identify the bacteria represented in DGGE gel. The products were subjected to PCR as templates with 341F (5′-CCTACGGGAGGCAGCA-3′, without GC-clamp) and 534R primers according to the following program: 3 min at 94°C, 30 cycles consisting of 15 s at 94°C, 20 s at 55°C, and 20 s at 72°C, and finally 5 min at 72°C. The PCR products were purified with a Universal DNA Purification kit (Tiangen, China) and cloned into E. coli TOP10 competent cells (Tiangen) with pMD 18-T Simple Vector (TaKaRa, China). The sequencing of plasmids with the desired DNA inserts was carried out by Biomed Biotech Co. Ltd. (China) using an M13F primer. The obtained sequences were compared with the sequences in the GenBank database with the BLASTn algorithm.
The data for TVCs were analyzed by the Duncan’s multiple comparison method at the significance level of 0.05 using IBM SPSS Statistics for Windows, Version 20.0 (IBM Corp., NY, USA). DGGE gel images were analyzed using Quantity One software (version 4.2, Bio-Rad) according to the user’s guide. The total intensity of all bands in each lane was defined as 100%, and relative intensities of each band in the same lane were calculated. Principal component analysis (PCA) was used to reduce the dimensions of the original variables to visualize the dynamics of the microbial community compositions over the chilled storage period with Canoco (Wageningen, Netherlands) for Windows package (Version 4.5).
Results
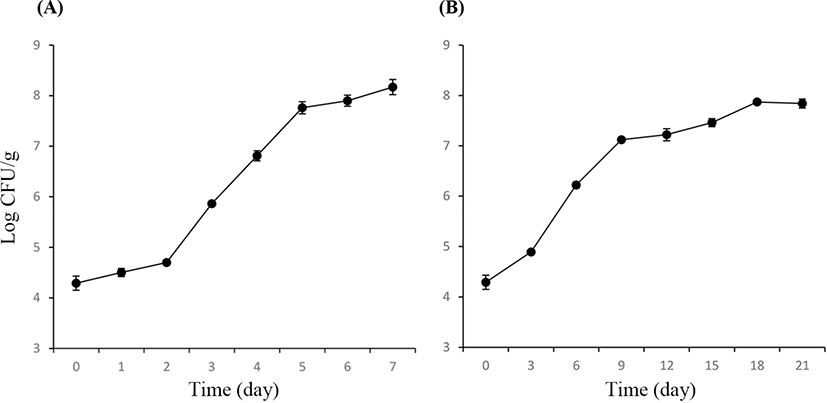
The TVCs of PT and VAC lamb are shown in Fig. 1. The initial TVC was 4.29±0.14 Log CFU/g. For the lamb in a PT, the TVC increased to more than 107 CFU/g on Day 5, exceeding 108 CFU/g on Day 6. For the lamb in VAC, the TVC increased to more than 107 CFU/g on Day 9 but was less than 108 CFU/g throughout the total process of storage (21 days). On Days 3 and 6, the lamb stored in a PT had significantly higher TVCs compared with the lamb stored in a VAC (p<0.05), indicating that the vacuum package could suppress the increase in TVCs more effectively than the air storage.
The bacterial communities of chilled lamb in a PT and a VAC were represented by the DGGE profiles, as shown in Fig. 2. A total of 15 bands were detected for the lamb in a PT and 16 bands for the lamb packaged in a VAC. All of the bands that appeared during the storage period were punched to identify the bacterial species. The sequencing results are shown in Tables 1 and 2 for the DGGE profile of lamb in a PT and a VAC, respectively. The dynamics of the microbial populations of lamb under different storage conditions are summarized in Table 3.
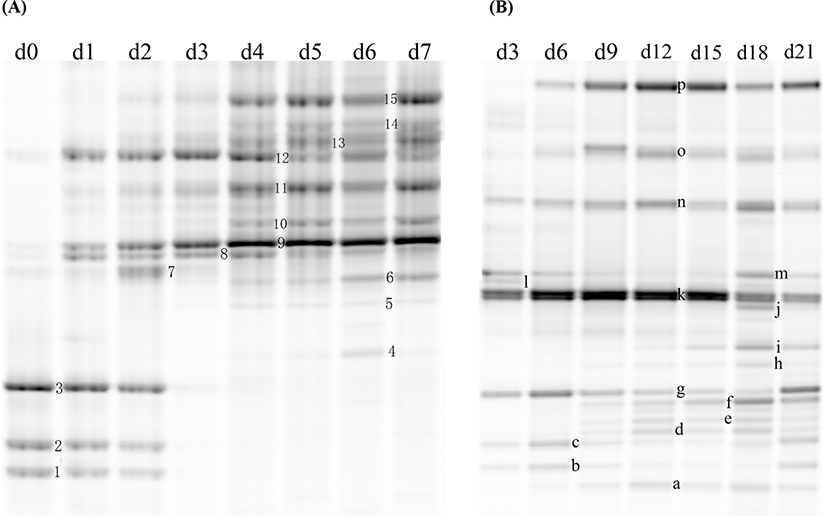
A total of 6 original microbial species (12 bands) were detected by DGGE in lamb meat, including Achromobacter spp. (Band 1), Acinetobacter spp. (Bands 9 and 10), Burkholderia cepacia (Bands 3 and g), Pseudomonas fragi (Bands 11, 12, and 13), Pseudomonas spp. (Bands 2 and c), and Psychrobacter fozii (Bands 8 and 1).
For the lamb in a PT, the intensity of Bands 1, 2, and 3, identified as Achromobacter spp, Pseudomonas spp, and Burkholderia cepacia, decreased during the storage and could not be detected by DGGE on Day 3. Bands 9 and 10, identified as Acinetobacter spp, increased from Day 1 and dominated in the latter period of storage with the relative abundance of more than 60%. Bands 11, 12, and 13, identified as different strains of P. fragi, appeared throughout the storage process, and their relative abundance increased from Day 1, peaked on Day 3, and then decreased gradually in the latter period of storage. Band 15, identified as P. fluorescens, increased from Day 2 to more than 10% after 5 days of storage, which was higher than that of P. fragi on Day 7. The Pseudomonas became dominant bacteria since Day 3, which the relative abundance of total Pseudomonas (sum of Pseudomonas fluorescens, Pseudomonas fragi and Pseudomonas spp.) was as high as 46.3% on Day 3 (Fig. 3). Bands 8 and 14 with low intensity during late storage period were identified as Psychrobacter fozii and Psychrobacter cibarius, respectively. Bands 4, 5, and 6 were identified as Serratia spp, Enterobacter spp, and Morganella spp, respectively, which belong to Enterobacteriaceae; all of these with relatively low abundance appeared in the latter period of storage. The relative abundance of Enterobacteriaceae increased since Day 4, and got as high as 4.6% on Day 6 (Fig. 3).
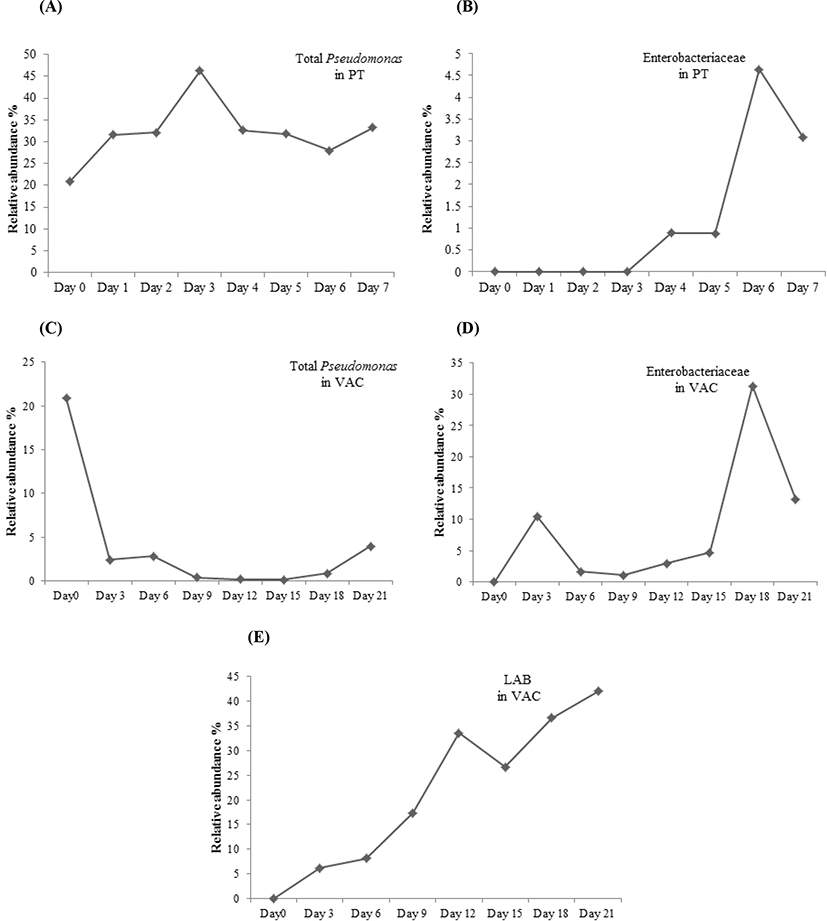
For the lamb packaged in a VAC, Band k dominated in the early period of storage, while its microbial species could not be identified (uncultured bacterium). Band g, identified as a strain of Burkholderia cepacia, appeared over the total storage period; its relative abundance showed a declining trend from Day 0 to Day 18 and then increased on Day 21. The dominant bacteria in the latter period of storage were LAB, consisting of Bands n, o, and p, which were identified as Carnobacterium divergens, C. maltaromaticum, and Lactococcus piscium, respectively. After 21 days of storage, LAB accounted for more than 40% of the total abundance (Fig. 3). Several Enterobacteriaceae strains, including Serratia spp. (Bands h and d), Serratia plymuthica (Bands b and e), Enterobacter spp. (Bands f and i), and Morganella spp. (Band m) were found in the lamb stored in a VAC. The relative abundance of Enterobacteriaceae was as high as 31.3% on Day 18 (Fig. 3). Morganella spp. appeared throughout the storage period, while the other Enterobacteriaceae strains appeared after 9 days of storage. Bands a, b, and c with low relative abundance were identified as Psychrobacter spp, Aeromonas salmonicida, and Pseudomonas spp, respectively, which appeared during the storage.
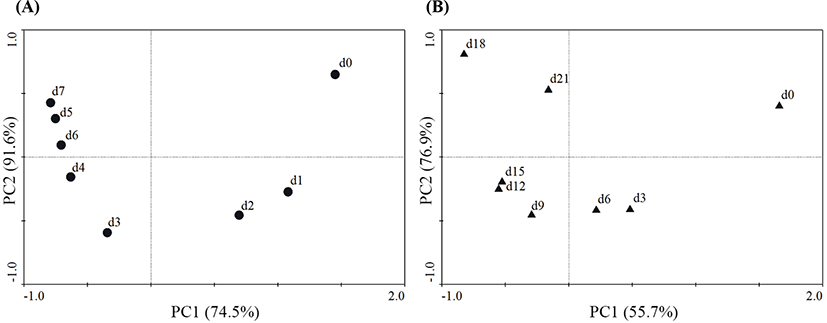
Additionally, changes in the microbial community compositions of the two groups at different sampling points were analyzed during chilled storage using PCA (Fig. 4). In Fig. 3A, PC1 and PC2 are 74.5% and 91.6%, respectively, of the total variation. InFig. 3B, PC1 and PC2 are 55.7% and 76.9%, respectively, of the total variation. For the lamb in a PT, the microbial community composition on Days 1 and 2 was quite different from that at the starting point (Day 0). The microbial community compositions were similar during the latter period of storage (from Day 3 to Day 7); however, they were largely different from the microbial community compositions of the early storage period (from Day 0 to Day 2). For the lamb packaged in a VAC, the microbial community compositions on Day 3 were quite different from those of Day 0, while the microbial community compositions were relatively similar during the middle period of storage (from Day 6 to Day 15). The microbial community compositions of the late storage period (Days 18 and 21) were largely different from those of other days, representing a similar situation as for the lamb stored in a PT. This showed that the microbial compositions of the lamb stored in a PT had faster dynamics than those of the lamb stored in a VAC.
Discussion
This study investigated the changes in the microbial communities of lamb packaged in a PT and a VAC during chilled storage. The microbial species were detected by PCR–DGGE technology to provide an overall picture of the microbial communities of lamb stored in a PT and a VAC over the total storage period. This study supplemented the bacterial communities of lamb meat from Chinese market, and provided dynamics of microflora of chilled lamb in the predominant packing form, the air-packed. The results represented basic data for improvement of hygiene and shelf life of the lamb meat. The lamb stored in a PT had higher viable counts than that packaged in a VAC. The TVCs of the lamb stored in a PT increased to more than 107 CFU/g, which was considered the spoilage level (Balamatsia et al., 2007; Stoops et al., 2015), on Day 5. However, they increased to more than 107 CFU/g on Day 9 for the lamb packaged in a VAC. This indicated that vacuum could inhibit the bacterial growth to prolong the shelf life of the lamb.
Meat spoilage is mainly caused by microbial metabolic activity, while only a subset of the initial microbial communities has been implicated in meat spoilage due to the effect of storage conditions, such as temperature and packaging environment (Casaburi et al., 2015). In this study, Pseudomonas spp. (Pseudomonas fragi and Pseudomonas fluorescens) and Acinetobacter spp. outcompeted the other bacteria and became the dominant bacteria in the lamb stored in a PT during chilled storage. Pseudomonas spp. and Acinetobacter spp. are aerobic bacteria; they dominate in lamb meat, causing spoilage with oxygen (Casaburi et al., 2015). For the lamb packaged in a VAC, LAB, consisting of C. divergens, C. maltaromaticum, and Lactococcus piscium, outcompeted the other microbial species and became the dominant bacteria in the latter period of storage. C. divergens, C. maltaromaticum, and Lactococcus piscium are facultative anaerobic bacteria that are able to adapt to the environment of vacuum. In addition, LAB can produce bacteria-inhibiting substances, such as organic acids and bacteriocins, which can prevent the growth of other spoilage-related bacteria (Cleveland et al., 2001; Ercolini et al., 2006; Wang et al., 2016).
Surprisingly, in a VAC, Pseudomonas spp. appeared over the total chilled storage; they are aerobic bacteria (Sun and Holley, 2012). Although Pseudomonas spp. appeared in the lamb packaged in a VAC, they were different from the strains (P. fragi and P. fluorescens) found in the lamb stored in a PT. As reported previously, although Pseudomonas spp. are aerobic, some strains can still grow in a micro-aerobic to an anaerobic environment. Also, some Pseudomonas strains are well adapted to thrive under such conditions and contain multiple enzyme systems for energy generation under oxygen-restricted or even anaerobic conditions (Schobert and Jahn, 2010). As reported by Soldatou et al. (2009), the number of Pseudomonas spp. increased to more than 3 Log CFU/g in a VAC, 30% CO2/70% N2, and 70% CO2/30% N2 packages without oxygen in 12 days. Further, in a previous study by Ercolini et al. (2011), the relative abundance of Pseudomonas spp. increased from 9.7% on Day 35% to 39.6% on Day 45 in a VAC. Therefore, the findings indicated that the number and relative abundance could still increase in the atmosphere with a low level of oxygen.
For the lamb stored in a PT, Enterobacteriaceae with relatively low abundance appeared in the latter period of storage, while it appeared in the lamb stored in a VAC throughout the storage period with an increasing abundance during the latter storage period. This indicated that Enterobacteriaceae could accelerate the meat spoilage, consistent with previous findings (Doulgeraki et al., 2012; Sun and Holley, 2012). Serratia, Morganella and Enterobacter were belong to Enterobacteriaceae family found in this study. They are facultatively anaerobic which are commonly found in vacuum packed meat product (Brightwell et al., 2007; Gill and Newton, 1979). The relative abundance of Enterobacteriaceae in the lamb stored in a VAC was much higher than that in the lamb stored in a PT during the total storage period, indicating a more important role of Enterobacteriaceae in the lamb packaged in a VAC than that stored in a PT.
Both the storage atmospheres could help detect Achromobacter, Pseudomonas and Burkholderia cepacia. These bacteria disappeared in the lamb stored in a PT after 3 days of storage. Burkholderia cepacia appeared over the whole storage period with a high relative abundance on Day 21 for the lamb packaged in a VAC. Achromobacter is close to Pseudomonas, while Burkholderia cepacia could produce lipase. The number of these bacteria generally increased during the storage. We speculated that disappearance of these bacteria in PT samples after Day 3 may be due to the increased abundance of other relative bacteria, such as Pseudomonas fluorescens and Enterobacteriaceae, which competition may contribute to this progress. Moreover, the role of these in the lamb meat spoilage and their ecological function in the meat remained unclear.
The microbial community compositions of lamb packaged under both the conditions in the late storage period were largely different from those in the early storage period, which was in agreement with the results of a previous study (Wang et al., 2016). This could be due to the competitive growth or inhibition between bacteria, resulting in the changes in the microbial compositions of lamb over the total storage period (Zhao et al., 2015). The microbial community compositions of lamb stored in a PT changed faster than those of lamb packaged in a VAC, indicating that vacuum packaging could delay the changes in the microbial community compositions. This might be due to higher microbial metabolism in lamb stored aerobically than that in a VAC. This could result in a change in the meat matrix, leading to a change in the microbial communities of lamb.
The feeding, slaughter environment, and contamination of carcass affect initial bacteria of lamb meat, and faeces and wool were major source of contamination. Ortuno et al. (2017) reported that dietary rosemary extract inhibited total viable and LAB and Enterobacteriaceae of lamb meat, which rosemary extract may reduced the shedding of these bacteria. The wool was considered as a important source of contamination for lamb carcass, and shearing and pre-slaughter washing significantly affected the bacterial loading of lamb meat (Biss and Hathaway, 1996; Hauge et al., 2011). Besides these factors, the breeds of sheep could also influence the microbial communities of lamb meat (Soldatou et al., 2009; Wang et al., 2016; Zhang et al., 2016). Therefore, it is necessary to analyze microflora of lamb meat from regional market. Compared with other meat like pork and beef, higher pH (≥5.8) and different nutritional composition could lead to specific bacterial communities in lamb meat (Mills et al., 2014). Previous study indicated that certain species of Enterobacteriaceae were able to grow aerobically/anaerobically on adipose and muscle tissues with high pH (>6), and were prevalent on lamb (Sun and Holley, 2012). Based on previous studies, Micrococcaceae were generally found in chilled pork, while Rahnella could be found in chilled beef (Pennacchia et al., 2011; Zhao et al., 2015). In this study, we observed Burkholderia cepacia in lamb, especially in VAC samples, which were rarely reported in lamb meat. Future study could focus on the regional characteristic of microflora in lamb meat.
In conclusion, the molecular biological method helped assess the changes in the microbiota of lamb stored in a PT and packaged in a VAC in detail. For the lamb stored in a PT, the main microbial species were P. fragi, P. fluorescens, and Acinetobacter spp. For the lamb packaged in a VAC, the main species were LAB comprising C. divergens, C. maltaromaticum, and Lactococcus piscium. The microbial compositions of lamb stored in a PT changed faster than those of lamb packaged in a VAC. The findings of this study may guide improve the lamb hygiene and prolong the shelf life of the lamb.













