Introduction
Listeriosis is a foodborne disease caused by Listeria monocytogenes. It can lead to sepsis, meningitis, encephalitis, and even death (de Noordhout et al., 2014). Despite its low incidence compared with that of other foodborne illnesses, listeriosis is one of the major issues in the food industry because of its high fatality rate. L. monocytogenes is found in dairy products, particularly in ready-to-eat cheese products. As L. monocytogenes survives in various environments such as a those with a wide range of temperature (0°C–45°C) and pH (4.1–9.6), it can contaminate cheese at several stages of production; therefore, its growth is difficult to control (Lungu et al., 2009; Melo et al., 2015).
Several methods have been used to control the growth of L. monocytogenes in cheese, including using bacteriocin or bacteriocin-producing lactic acid bacteria (LAB). Bacteriocins are peptides or proteins, ribosomally synthesized by bacteria, which have antimicrobial ability against closely related species. The application of bacteriocin-producing bacteria is advantageous as they are stable, cost-effective, and safe. Anti-listerial activity of LAB in cheese have also been reported (Coelho et al., 2014; Dal Bello et al., 2012; Kondrotiene et al., 2018).
In the present study, we aimed to determine the effects of bacteriocin-producing LAB isolated from raw milk on the growth of L. monocytogenes in milk broth and cheese.
Materials and Methods
Potential bacteriocin-producing LAB were isolated from raw bovine milk, obtained from a Chung-Ang University-affiliated farm (Anseong, Korea). The sample was serially diluted ten-fold and plated on MRS agar (BD Difco, Franklin Lakes, NJ, USA). The plates were incubated at 37°C for 24–48 h, and a total of 90 well-isolated colonies were collected. Each colony was inoculated into MRS broth for 24 h at 37°C.
To screen for antimicrobial activity, the cell-free supernatant (CFS) was obtained after neutralization with 1N NaOH, centrifugation at 15,000×g for 10 min at 4°C, and filtered through 0.45 μm filters to remove bacterial cells. Then, each supernatant was spotted on the tryptic soy agar (TSA; BD Difco) plate inoculated with a lawn of L. monocytogenes ATCC 19115 as an indicator strain. The plates were incubated at 30°C for 12 h, and antibacterial activity was confirmed with the presence of inhibition zone. The strains with antibacterial activity were routinely cultured in MRS broth at 37°C overnight and were preserved in 10% skim milk supplemented with 25% (v/v) glycerol, stored at –80°C for further use.
The bacteriocin-producing strains were identified by Gram staining, carbohydrate fermentation profile [analytical profile index (API) test], and 16S rRNA gene sequencing analysis. Gram staining and API analysis was performed using a Gram-stain kit (BD Difco) and API 50 CHL kit (bioMérieux, Marcy-l’Étoile, France), respectively, according to the manufacturer’s instructions.
For 16S rRNA analysis, the genomic DNA was extracted using QIAamp PowerFecal DNA Kit (Qiagen, Hilden, Germany) and amplified using 2X H-star Taq polymerase chain reaction (PCR) Master Mix (BioFACT, Daejeon, Korea). PCR was performed using the universal bacterial primers 27F (5′-AGAGTTTGATCMTGG CTCAG-3′), 1492R (5′-TACGGYTACC TTGTTACGACTT-3′), 785F (5′-GGATTAGA TACCCTGGTA-3′), and 805R (5′-GACTACCAGGGTATCTAATC-3′). The PCR products were purified using a PCR purification kit (Qiagen) and sequenced by SolGent (Daejon, Korea).
The analyzed sequences were confirmed using the EzTaxon-e server (https://www.ezbiocloud.net/; Kim et al., 2012) and NCBI GenBank database using the Basic Local Alignment Search Tool (BLAST) algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1990).
Bacteriocin activity was assessed using a spot-on-lawn method as described previously (Phumisantiphong et al., 2017) with minor modifications. Briefly, each indicator strain was inoculated to 4 mL of molten TSA and overlaid on the base TSA plate. After solidification, 20 μL of the neutralized CFS of LAB strains was spotted onto the indicator lawn. After incubation at 30°C for 12 h, a clear inhibition zone was observed. The foodborne pathogens used as indicator strains were cultured in tryptic soy broth (TSB; BD Difco) at 37°C overnight before use. The experiment was conducted in triplicates.
The acid production of the Lactococcus lactis CAU2013 was evaluated and compared with that of the commercial starter TCC-3 (Chr. Hansen, Hørsholm, Denmark), which consisted of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus. The cultures were grown in MRS broth at 37°C overnight. Then, individual cultures and mixture of L. lactis CAU2013 and TCC-3 (1:1 ratio) were inoculated in 10% skim milk broth and whole milk. The pH and titratable acidity (TA) were measured every three hours for 12 h while incubating at 30°C. To determine TA, 0.1% phenolphthalein was used as an indicator, and 0.1 N sodium hydroxide (NaOH) for titration.
To determine the anti-listerial properties of strain CAU2013 when used as an adjunct starter in milk, 10% skim milk broth and whole milk media were inoculated with an overnight culture of CAU2013 and 1:1 ratio of CAU2013 and TCC-3 starter (final concentration of 7 Log CFU/mL). Additionally, the overnight culture of L. monocytogenes ATCC 19115 was inoculated to each setup (final concentration of 5 Log CFU/mL). The inoculated milk media were incubated at 30°C for 12 h. The viable cell count of L. monocytogenes was determined every three hours. Samples were diluted serially in ten-fold increments using 1×phosphate-buffered saline (PBS; pH 7.5) and plated on Oxford agar (BD Difco).
The lab-scale cheese was manufactured following the methods of Mills et al. (2011) with some modifications. TCC-3 was used as the primary starter and L. lactis CAU2013 as an adjunct culture. The starter cultures were initially grown in MRS broth at 37°C for 24 h before inoculation into 10% skim milk broth and incubated for 18 h at 37°C before use. Additionally, L. monocytogenes ATCC19115 was cultured in TSB for 18 h at 37°C before use.
Milk (400 mL; Seoul Milk, Seoul, Korea) was heated to 31°C before the inoculation of starter culture. The starter cultures were inoculated as follows: TCC-3 and L. lactis CAU2013 and TCC-3 (1:1 ratio), both at a final concentration of 7 Log CFU/mL. Subsequently, 0.01% L. monocytogenes at a level of 5 Log CFU/mL was inoculated into both treatments. After 30 min, 0.2 g/L of rennet was added, and the mixture was stirred for 2 min. Once coagulum formed firmly, the curd was cut into cubes, and the mixture was stirred for 10 min. Then, the mixture was heated to 36°C for 10 min and stirred for 20 min. The whey was drained off, and curd was distributed into the sterile dish. The samples were stored at 4°C and 10°C for 5 days. The procedure of cheese production is illustrated in Fig. 1.
The viable cell counts of LAB and L. monocytogenes in the lab-produced cheese were determined in duplicate every day during storage at 4°C and 10°C. For microbial analysis, 1 g of cheese was homogenized in 9 mL of PBS buffer and were serially diluted ten-fold in the same buffer and plated on the appropriate agar plate. The LABs were enumerated on MRS agar after incubation at 37°C for 3 days, and L. monocytogenes on Oxford agar after incubation at 37°C for 24 h. All of the experiments were conducted in triplicates.
Results and Discussion
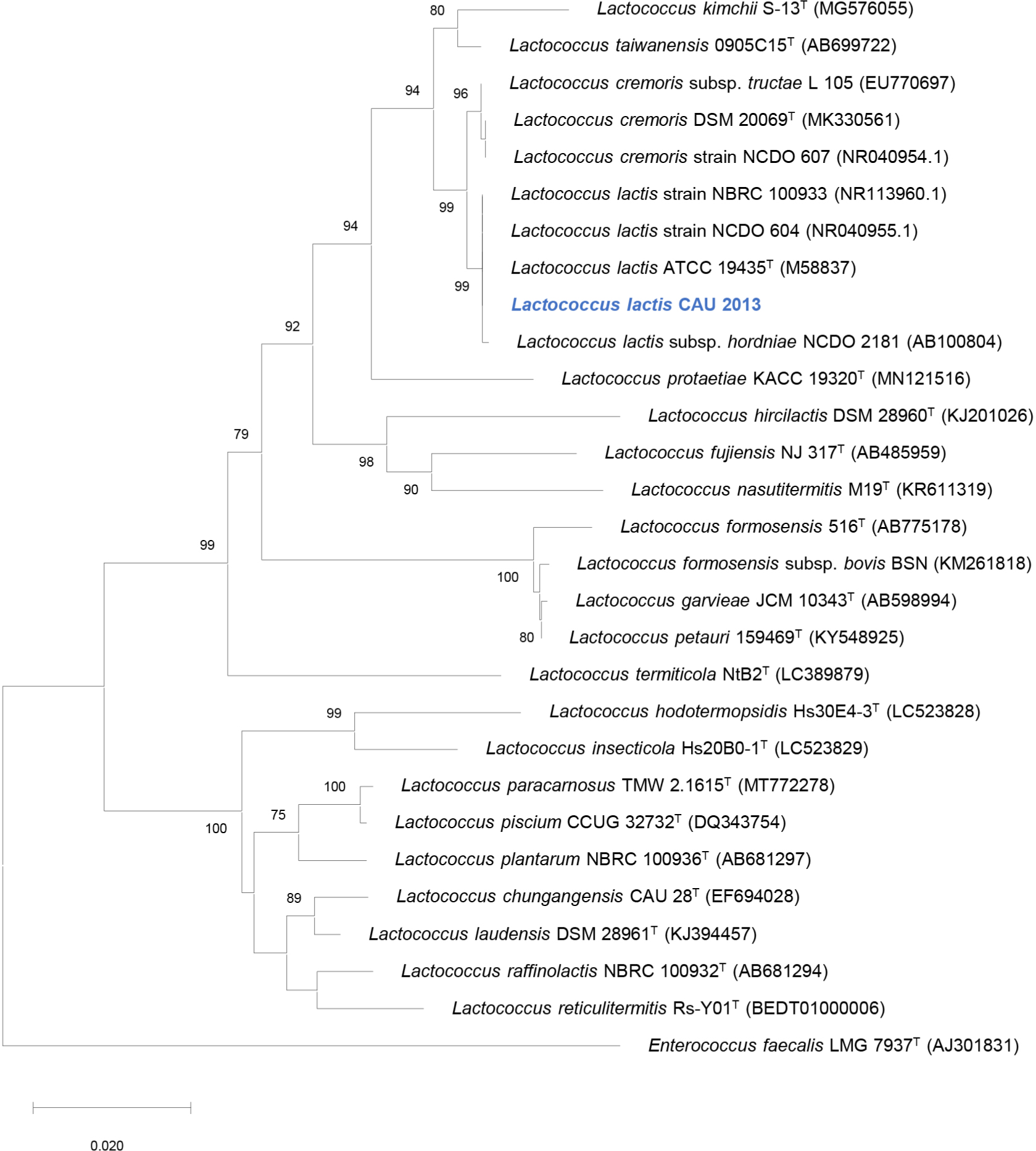
Among the 90 colonies isolated from raw milk, one isolate exhibited antibacterial activity against L. monocytogenes. The strain CAU2013 was characterized as a gram-positive, coccus-shaped bacterium. The biochemical characteristics determined using the API 50 CHL kit are described in Table 1. 16S rRNA gene sequence analysis revealed that strain CAU2013 is most likely a strain of L. lactis (Table 2), which commonly produce nisin (Shin et al., 2016). Neighbor-joining phylogenetic tree of the strain CAU 2013 and related type strains based on 16S rRNA gene sequences also clearly show that this strain belongs to L.lactis (Fig. 2). L. lactis strains are historically used in the fermentation and preservation of food and are generally recognized as safe (Cook et al., 2018). Therefore, L. lactis CAU2013 was selected for downstream applications in the study.
L. monocytogenes ATCC 19115 was used as an indicator strain for all experiments because it belongs to the serotype 4b, which causes most cases of listeriosis.
The bacteriocin produced by L. lactis CAU2013 had antibacterial activity against all Listeria strains as well as Staphylococcus aureus, which are common foodborne pathogens (Yoon, 2020). However, no antibacterial activity was observed against other gram-positive foodborne pathogens, such as Salmonella enteritidis and Escherichia coli (Table 3). Generally, nisin is highly effective against gram-positive bacteria by binding to lipid °C, which leads to the inhibition of cell wall biosynthesis or pore formation in the membrane. However, nisin cannot bind to its target lipid II in gram-negative bacteria, because of the presence of the outer membrane (Li et al., 2018).
The changes in pH and TA values in 10% skim milk broth and in whole milk are presented in Fig. 3. L. lactis CAU2013 reduced the pH of skim milk broth from 6.41 to 5.77 and that of whole milk from 6.65 to 6.20. Additionally, TA value increased to 0.25 in both broths. Ayad et al. (2004) described fast, medium, or slow-acidifying strains as ΔpH (=pHat time–pHzero time) of 0.4 U achieved after 3 h, 3–5 h, and >5 h, respectively. Also, Raquib et al. (2003) classified strains with TA as low, moderate, or fast when the TA values were <0.5, between 0.5 and 0.6, and >0.6, respectively. Therefore, L. lactis CAU2013 can be classified as a low acidifier strain. This result is consistent with other studies that reported poor acid production from L. lactis strains (Ayad et al., 2004; Coelho et al., 2014).
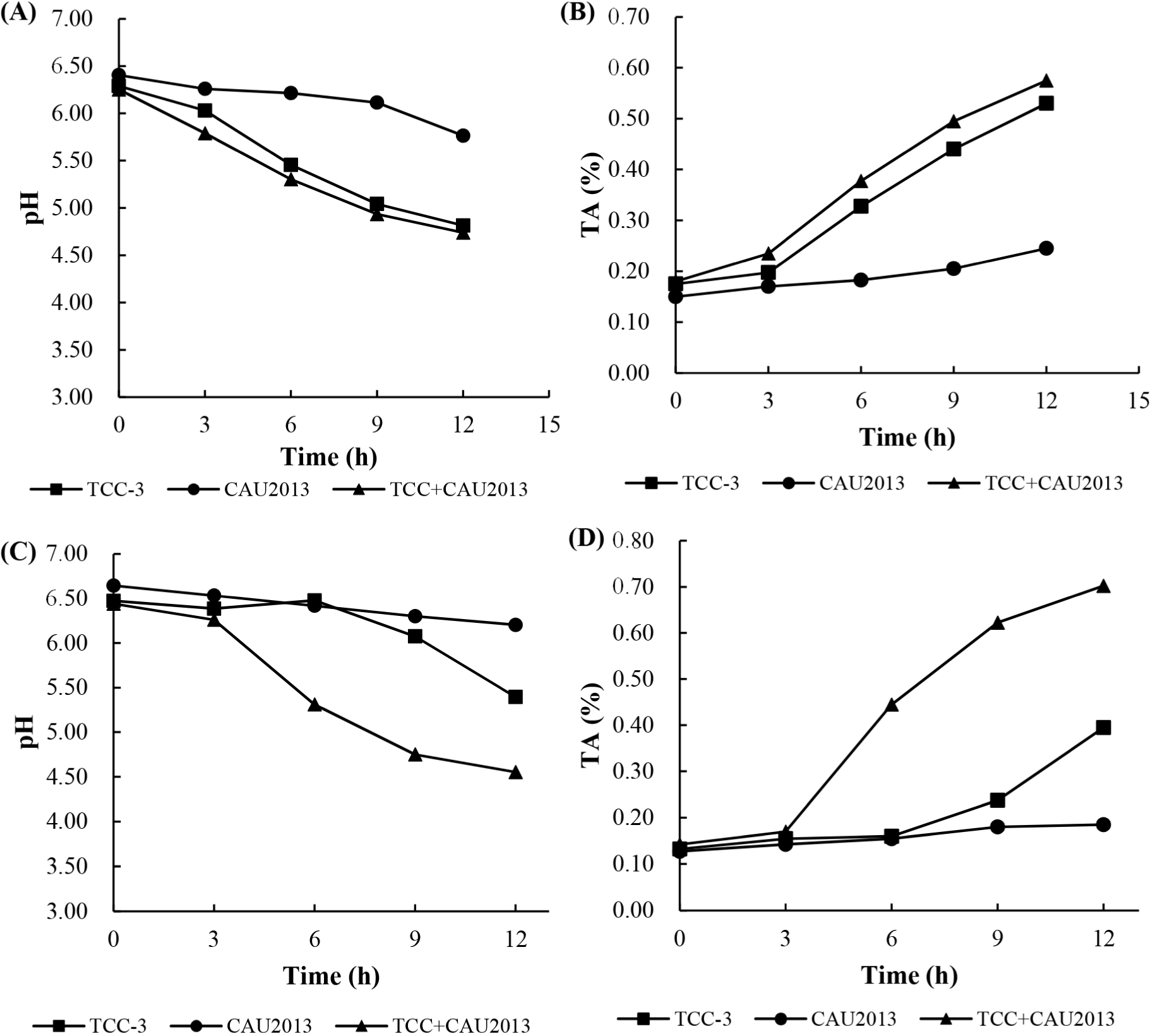
The pH values measured corresponded with the calculated TA and were generally similar for skim milk broth and whole milk. The mixed starter, consisting of TCC-3 and L. lactis CAU2013, accelerated the acidification in milk. Nevertheless, bacteriocin-producing strains delay acidification (Garde et al., 1997); however, the strain CAU2013 did not show similar properties. The accelerated acidification might be because of the interaction between the strains; however, the underlying mechanisms need further research. Ávila et al. (2005) observed that enterocin-producing adjunct starter enterococci enhanced milk acidification, which may be stimulated by the low-molecular-weight nitrogen compounds produced by primary starter, Lactobacillus helveticus LH92.
The rapid decline in pH during the initial stage of cheese production is crucial for curd formation and prevention of the growth of undesirable microorganisms. Therefore, the fast-acidifying strains can be used as primary starters, while the slow-acidifying bacteria can be used as adjunct starters. As the strain CAU2013 has antibacterial property but has low acid production ability, it is better suited as an adjunct starter culture.
The growth of L. monocytogenes was monitored in skim milk broth and whole milk during incubation at 30°C. In skim milk broth with L. lactis CAU2013, the concentration of L. monocytogenes count was reduced by 3 Log units more compared with that of other samples after 3 h and not detected following 6 h of fermentation (Fig. 4A). In the whole milk with the strain CAU2013, L. monocytogenes count was reduced by 0.5 Log unit after 6 h, and 1 Log unit after 9 h compared with that of other samples (Fig. 4B).
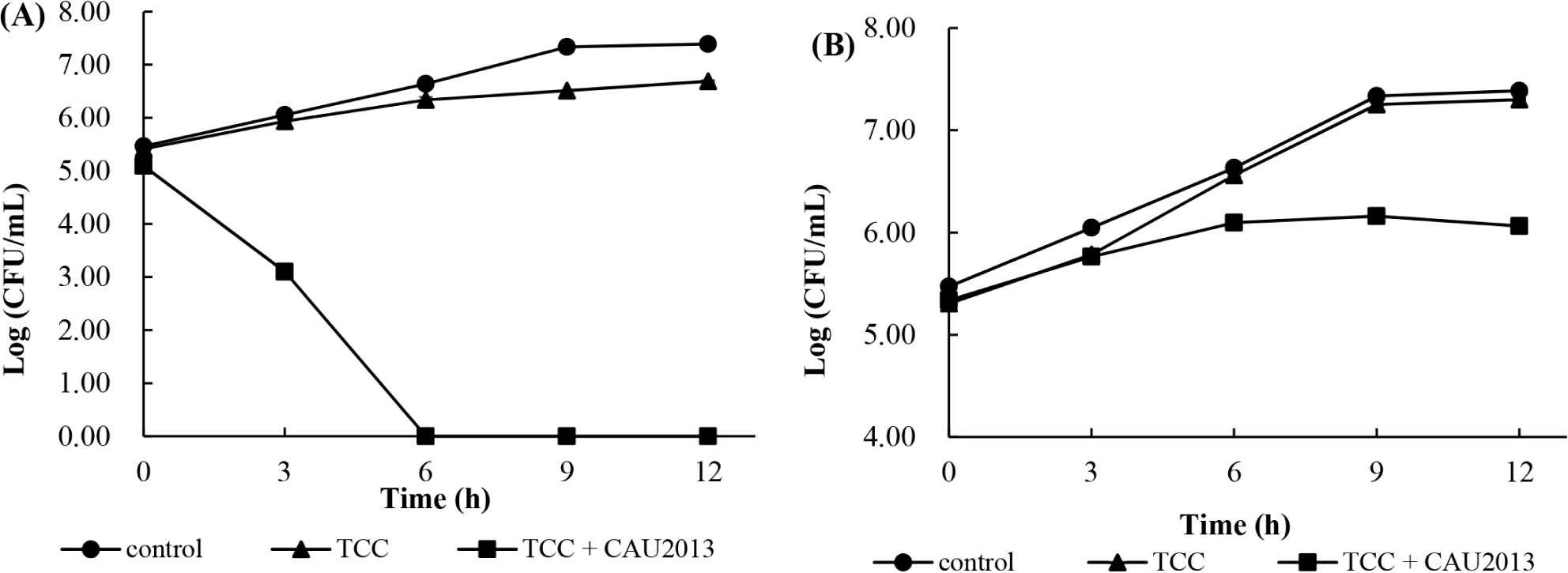
The results support the findings from several studies that reported that the addition of bacteriocin affects the biocontrol of spoilage bacteria. Muñoz et al. (2007) investigated that E. faecalis-produced enterocin in milk and found that it could control the growth of Staphylococcus aureus. In addition, according to Arqués et al. (2011), the addition of nisin in milk decreased L. monocytogenes count by 3 Log units after 4 h.
The efficiency of the combined starter cultures in the inhibition of L. monocytogenes was lower in whole milk than in skim milk. The difference in the composition between the two milk media could be a factor responsible for the difference. In addition, Muñoz et al. (2007) stated that low effectiveness in foods could be attributed to higher retention of the bacteriocin molecules by milk components, resulting in slower diffusion. However, in both cases, inhibition of L. monocytogenes growth was observed. The results suggest the potential application of L. lactis CAU2013 in various food systems to control L. monocytogenes growth.
The cell count of the starter cultures was determined during the storage at 4°C and 10°C (Fig. 5). In both the cases, LAB reached a final concentration of 9 Log CFU/g during cheese manufacture.
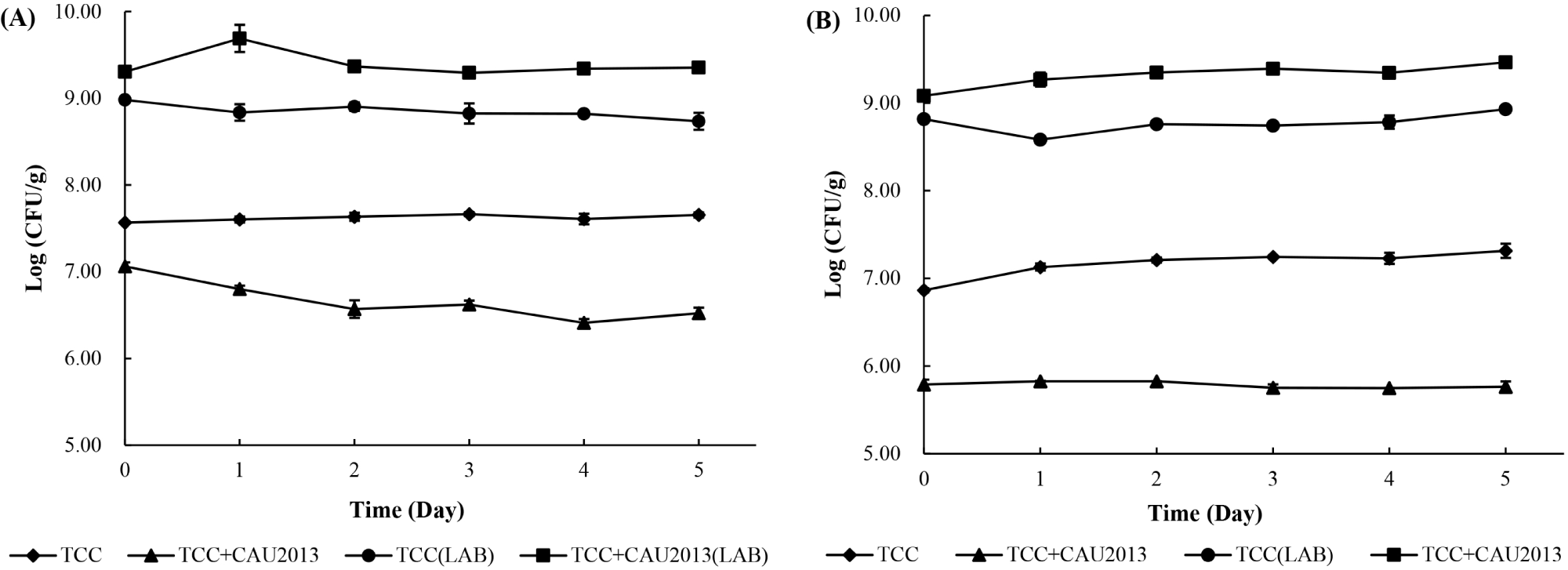
During the storage at 4°C, the cheese treated with TCC-3 starter culture maintained L. monocytogenes count at 7.5 to 7.7 Log CFU/g. In contrast, the cheese treated with TCC-3 and L. lactis CAU2013 had less L. monocytogenes count, approximately 0.5 Log unit at 0 h and 1 Log unit after 5 days with a final concentration of 6.4 Log CFU/g. Besides, during the storage at 10°C, the cheese treated with TCC-3 starter culture maintained the bacterial count between 6.86 and 7.31 Log CFU/g (Fig. 5A). within contrast, the cheese treated with TCC-3 and CAU2013 had less L. monocytogenes count, approximately 1 Log unit at 0 h and 1.5 Log unit after 5 days, with a final concentration of 5.76 Log CFU/g (Fig. 5B). This result is consistent with a study that reported that 2 Log unit reduction was observed in cheese with L. lactis strain (Coelho et al., 2014). Moreover, Kondrotiene et al. (2018) showed that nisin-producing L. lactis strains decreased the growth of L. monocytogenes in fresh cheese during 7 days of storage at 4°C.
Therefore, the results support that manufacturing cheese using a bacteriocin-producing starter reinforced the inhibition of growth of L. monocytogenes, and it would be effective in controlling contamination during cheese production. Additionally, after storage at temperatures of 4°C and 10°C, L. monocytogenes count was reduced, which may confirm the potential of LAB in controlling the growth of L. monocytogenes during storage at refrigeration temperature.














