Introduction
Listeria monocytogenes is a Gram-positive pathogenic bacterium found in humans and animals (McLauchlin et al., 2004; Yoon and Kim, 2022). It has a high infection ability owing to host-pathogen interactions (Tsai and Chen, 2020). In humans, it is mainly transmitted through food and can cause listeriosis, which is especially fatal to children, elderly people, and immune-deficient patients (Gandhi and Chikindas, 2007). Listeriosis is caused by consuming food contaminated with L. monocytogenes, such as non-pasteurized milk, cheese, undercooked meat, and smoked ducks (Aygun and Pehlivanlar, 2006; Jang et al., 2021; Park et al., 2021). In particular, its ability to grow at low temperatures, even refrigerated temperatures, prevents its use in human foodstuffs (Possas et al., 2022).
Many biological agents that prevent contamination by or inhibit the growth of L. monocytogenes have been studied, and bacteriocins are essential candidates for controlling L. monocytogenes (Perumal et al., 2019). Bacteriocins are ribosomally synthesized proteinaceous substances produced by bacteria that are active against other bacteria (Cotter et al., 2005; Lee et al., 2021) and have been proposed as an alternative to antibiotics to inhibit the growth of pathogenic microorganisms (Cotter et al., 2013). Bacteriocins with antilisterial activity have been isolated from diverse lactic acid bacteria (LAB) such as Enterococcus spp., Lactobacillus spp., Leuconostoc spp., and Weissella spp. (Dong et al., 2022; Ghalfi et al., 2006; Todorov, 2010; Woo et al., 2021; Yang and Moon, 2021). Among them, nisin, an antibiotic bacteriocin produced by Lactococcus lactis, is the only bacteriocin used in the food industry in many countries to improve food safety and extend the shelf life of foods (Churklam et al., 2020; van Kuijk et al., 2012). However, nisin-resistant L. monocytogenes mutants have also been reported (Sabaté and Audisio, 2013; Vadyvaloo et al., 2004), suggesting that new biological agents should be developed to overcome nisin resistance.
Ligilactobacillus agilis is a motile lactic acid bacterium generally isolated from the gastrointestinal tract of mammals (Baele et al., 2001; Kajikawa et al., 2018). Several L. agilis have been shown to inhibit L. monocytogenes (Gudiña et al., 2015; Shi et al., 2021). However, more information must be given regarding the antilisterial substances from L. agilis. Here, we report the isolation of L. agilis C7, which inhibits L. monocytogenes that contaminates various foods including meat and dairy products, and characterize a novel bacteriocin from L. agilis C7, named agilicin C7, at the genetic and molecular levels by heterologous expression in Escherichia coli.
Materials and Methods
Feces from healthy pigs were used to isolate LAB with antilisterial activities. One gram of fecal sample was mixed with 30 mL of sterile phosphate buffer (pH 7.4) and diluted ten-fold. The dilutions were spread on the de Man, Rogosa, and Sharpe (MRS; BD, Franklin Lakes, NJ, USA) agar plates containing 0.02% (w/v) sodium azide (Sigma-Aldrich, St. Louis, MO, USA) and 0.02% (w/v) Bromocresol Purple (BCP; Sigma-Aldrich). The plates were incubated at 37°C overnight. Colonies were selected and streaked onto MRS agar. To screen for anti-listerial LAB, the isolates were cultured in MRS broth at 37°C for 24 h, and the cell-free supernatant (CFS) was collected by centrifugation (8,000×g, 10 min, 4°C). The CFS was adjusted to pH 7.0 and filtered using a 0.45 μm pore size syringe filter (Minisart®, Sartorius, Göttingen, Germany). The antilisterial activity of CFS was measured using an agar well diffusion assay, with L. monocytogenes ATCC 19114 as an indicator strain (Yoon and Kim, 2022).
The 16S rRNA gene sequencing was used to identify the isolate with antilisterial activity. The 16S rRNA gene was amplified using a PCR Thermal Cycler (Dice® Gradient, TaKaRa, Shiga, Japan) with primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGTTACCTTGTTACGACTT-3’). The PCR condition was 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 1 min. A homology search was performed using the National Center for Biotechnology Information BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The kinetics of bacterial growth and production of antimicrobial substances by L. agilis C7 inhibiting growth of L. monocytogenes were investigated during batch fermentation. A culture of L. agilis C7 was inoculated at a 2% volume into 200 mL MRS broth and incubated at 37°C for 72 h. Samples were collected at designated times over a 72 h period. Cell growth was determined by measuring the optical density at 600 nm using a spectrophotometer (Shimadzu, Kyoto, Japan). The production of anti-listerial substances was compared by measuring the size of the inhibition zone of the neutralized CFS against L. monocytogenes.
Bioinformatic analyses of the L. agilis C7 genome, previously reported by Yoo et al. (2022), were conducted for mining for the bacteriocin gene, which is responsible for the anti-listerial activity, using BAGLE4 (http://bagel4.molgenrug.nl/), BACTIBASE (https://bactibase.hammamilab.org), AntiSMASH (https://antismash.secondarymetabolites.org), and BLASTP (https://blast.ncbi.nlm.nih.gov). Signal P 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) and MultAlin (http:// multalin.toulouse=ra.fr/multalin/) were used to predict signal peptides and for multiple sequence alignment with other bacteriocins, respectively.
Sequence analysis of the putative bacteriocin biosynthesis gene, agilicin C7, showed an Ala-Ser-Ala cleavage site at the N-terminus of the signal sequence (Biet et al., 1998; Worobo et al., 1995). Forward and reverse primers targeting the mature form of agilicin C7 were designed based on L. agilis C7 genome sequences (Yoo et al., 2022). PCR amplification was performed using forward (5’-AAA GAATTC CATATG AACTCGGGTTACTGTAAACC-3’) and reverse (5’-AAA GTCGAC CCCCCT ATAACCAGGTTG-3’) primers, and the PCR products were purified using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Düren, Germany). After the purified PCR products and pET21b vector were digested with NdeI and SalI for 1 h at 37°C, ligation was carried out using T4 Ligase overnight at 16°C. The pET-21b expression vector containing the agilicin C7 gene, named pET-21b-agiC7, was transformed into E. coli BL21 (DE3; Hwang et al., 2018). Then, recombinant transformants were selected on the Luria-Bertani (LB, BD) agar containing ampicillin (100 μg/mL).
Overnight culture of E. coli BL21 (DE3) harboring the plasmid pET-21b-agiC7 inoculated 1% (v/v) to LB broth containing ampicillin (100 μg/mL) and then was grown at 37°C. After inducing protein overexpression by adding 1 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG; Sigma-Aldrich), the cells were recovered by centrifugation at 10,000×g for 10 min. The cells were then resuspended in TN buffer (20 mM Tris-Cl, 100 mM NaCl, pH 8.0) and disrupted using a sonicator (VCX750; Sonics, Oklahoma City, OK, USA). The insoluble cytoplasmic fraction and cell debris were collected by centrifugation (13,000×g, 10 min, 4°C) because recombinant agilicin C7 (r-agilicin C7) was expressed as an inclusion body (data not shown), and the resulting pellet was dissolved in TN buffer containing 8 M urea. The His-tagged r-agilicin C7 was purified using nickel-nitroloacetic acid (Ni-NTA) affinity chromatography and analyzed by Tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram.
The purified r-agilicin C7 was incubated for 2 h at 37°C after adding proteinase-K, α-amylase, α-chymotrypsin, pepsin, and lipase at a 5 mg/mL concentration, respectively (Yang and Moon, 2021). All the enzymes were purchased from Sigma-Aldrich. After boiling for 5 min at 80°C to inactivate enzymes, the anti-listerial activity of the samples was evaluated by the agar well diffusion method (Yoon and Kim, 2022).
To examine the stability of the anti-listerial activity of r-agilicin C7 over a wide range of pH, the pH of r-agilicin C7-containing solutions was adjusted to pH 1.0 to 10.0 using sterile 1 N NaOH or 1 N HCl. After incubation at 37°C for 2 h, the samples were readjusted to pH 8.0 and tested using the agar well diffusion method (Hwang et al., 2018).
The effect of temperature on anti-listerial activity was tested by incubating at 4°C, 25°C, 37°C, 50°C, 60°C, 80°C, 100°C, and 121°C for 15 min. The treated samples were then cooled to room temperature, and their anti-listerial activity was evaluated using the agar well diffusion method (Hwang et al., 2018).
The effect of organic solvents on the anti-listerial activity of r-agilicin C7 was evaluated by adding 10% (final concentration, v/v) of acetone, acetonitrile, methanol, ethanol, chloroform, and isopropanol, respectively. After incubating at 37°C for 2 h, the anti-listerial activity of the samples was evaluated by the agar well diffusion method (Hwang et al., 2018).
The antimicrobial activity of r-agilicin C7 against indicator strains, including Gram-negative and Gram-positive bacteria, and 2 fungi (Table 1), was examined. After overlaying the soft agar (0.75% agar) containing indicator strains on agar media, the wells were punched into the agar plates using a sterile glass cylinder (Hwang et al., 2018). The wells were filled with 200 μL of r-agilicin C7 solution and incubated at the optimal growth temperature. The antimicrobial activity was measured based on a clear zone around the wells.
Antimicrobial activity was measured by agar well diffusion. Inhibition zone was expressed in degrees: −, no inhibition; +, up to 1 mm; ++, up to 5 mm.
KCCM, Korean Culture Center of Microorganisms; NB, nutrient broth (BD, Franklin Lakes, NJ, USA); KVCC, Korea Veterinary Culture Collection; LB, Luria-Bertani (BD); ATCC, American Type Culture Collection; BHI, brain heart infusion (BD); KCTC, Korean Collection for Type Culture; KFRI, Kerala Forest Research Institution; MRS, de Man Rogosa and Sharpe (BD); PDA, potato dextrose agar (BD); YM, yeast alt (BD).
Tricine SDS-PAGE was performed to confirm the molecular weight of the r-agilicin C7, as described by Hwang et al. (2018), using 4% acrylamide stacking gel, 10% acrylamide spacer gel, and 16.5% acrylamide separation gel. The samples were mixed with Tricine sample buffer (Bio-Rad Laboratories, Hercules, CA, USA). Electrophoresis was performed in an electrophoresis chamber (PowerPac 300; Bio-Rad Laboratories) at 30 V for 9 h. After running, one gel was stained with staining buffer (10% acetic acid, 0.025% Coomassie Brilliant Blue G-250 (Bio-Rad Laboratories), 45% methanol, and 45% distilled water) for 1 h and then destained with destaining buffer (10% acetic acid, 45% methanol, and 45% distilled water) for 12 h. The other gel was fixed with r-agilicin protein in a destaining buffer overnight and then washed in distilled water for 3 h. The washed gels were placed in the square dishes and overlaid with 0.75% (w/v) soft agar inoculated with L. monocytogenens ATCC 19114 (approximately 105 CFU/mL). The plate was incubated for 6 h at 37°C, and the gel was examined for the inhibition zone.
FE-SEM was used to observe morphological changes in L. monocytogenes ACTC 19114 treated with various concentrations of r-agilicin C7 in BHI broth. An overnight culture of L. monocytogenes ACTC 19114 (approx. 104 CFU/mL) was inoculated into BHI broth containing r-agilicin C7 (0, 160 AU/mL, 320 AU/mL, respectively) and incubated for 2 h at 37°C. The treated cells were collected by centrifugation at 6,000×g for 10 min, and the pellets were washed twice with 1 mM PBS buffer (pH 7.0). The cells were treated with 0.1 M sodium cacodylate buffer containing 4% glutaraldehyde (EMS, Hatfield, PA, USA) and washed with 0.05 M sodium cacodylate three times for 10 min. Post-fixation was carried out with 1% osmium tetroxide (EMS) and then washed with 0.05 M sodium cacodylate, as mentioned above. The fixed cells were resuspended in ethanol (50%, 75%, 90%, 95%, and 100% for 20 min each) and treated with 100% isoamyl acetate (Tokyo Chemical Industry, Tokyo, Japan) to dehydrate. The morphology of L. monocytogenes ATCC 19114 cells treated with r-agilicin C7 was observed using a SIGMA 500 FE-SEM (Carl Zeiss, Oberkochen, Germany).
Results
Bacterial strains with a yellow halo on the BCP-MRS medium were isolated from pig fecal samples. Their CFSs were screened for antimicrobial activity against L. monocytogenes ATCC 19114 using an agar well diffusion assay. The isolate with the strongest inhibitory activity was selected for further experiments. 16S rRNA gene sequencing showed a close phylogenetic similarity to L. agilis LGM7-7 (99.93%), then it was identified as an L. agilis species and designated L. agilis C7. The genome sequence of the L. agilis C7 strain was deposited in the GenBank database under accession no. JAMGEC000000000 (Yoo et al., 2022).
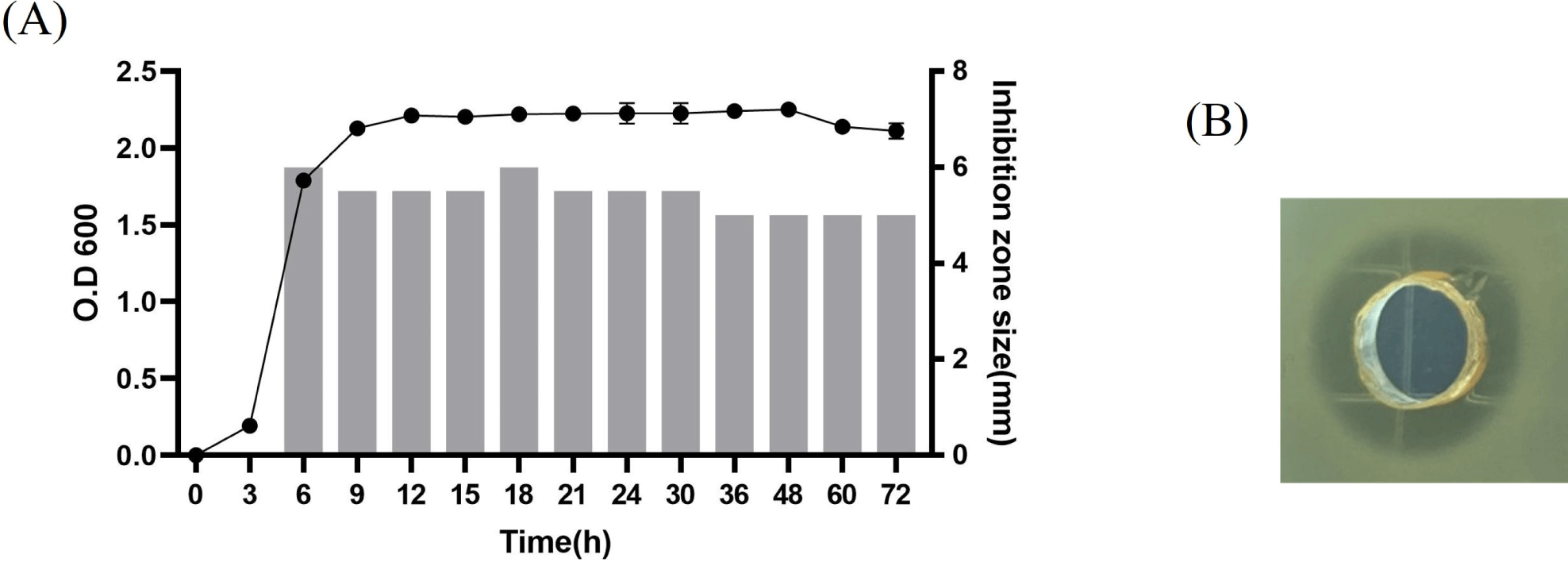
The kinetics of anti-listerial substance production in L. agilis C7 indicated that it was associated with cell growth, as shown in Fig. 1. The anti-listerial substance was detected in the logarithmic growth phase, and its activity was stably maintained during the stationary growth phase.
To unravel the genetic traits relevant to the anti-listerial activity of the C7 strain, bioinformatic analyses of the C7 genome were performed, which revealed a complete operon highly homologous to an operon for garvicin Q biosynthesis. It was selected for further investigation because Garvicin Q family class II bacteriocins inhibit L. monocytogenes (Tosukhowong et al., 2012). Fig. 2A shows the organization of the C7 gene cluster, which is highly homologous to the Garvicin Q bacteriocin. It consists of four genes encoding, ABP transporter permease, histidine kinase, bacteriocin (named “agilicin C7”), and bacteriocin immunity protein. The nucleotide and deduced amino acid sequences of agilicin C7 are shown in Fig. 2B. Amino acid sequence analysis of agilicin C7 revealed an Ala-Ser-Ala cleavage site at the N-terminus of its signal sequence, which is also found in several class IId bacteriocins (Worobo et al., 1995). The Ala-Ser-Ala site at the N-terminus of divergicin A serves as a signal peptide for the cell’s general export system, such as the E. coli sec pathway (Biet et al., 1998).
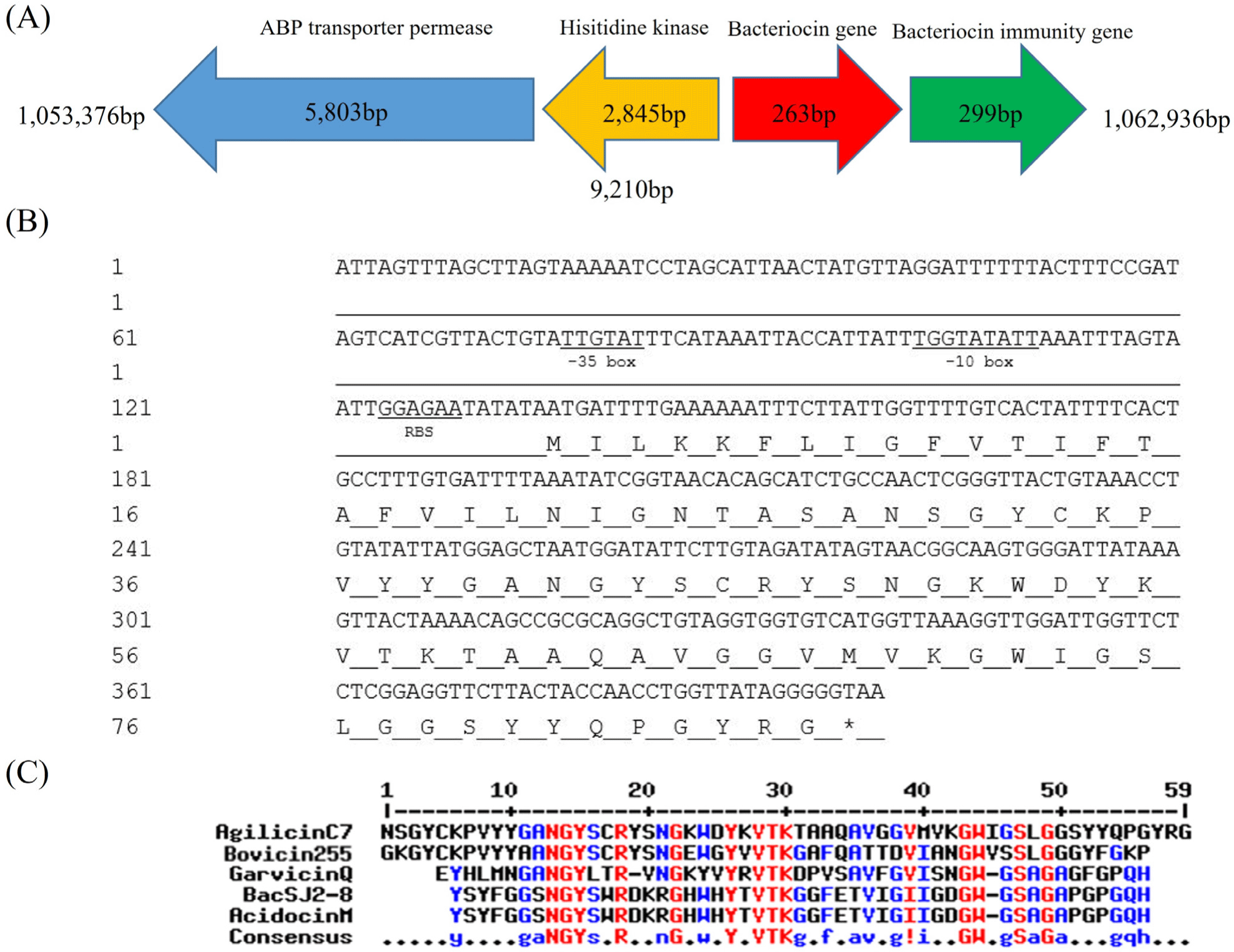
The putative amino acid sequence of agilicin C7 showed high homology with those of Class IId bacteriocins (Fig. 2C). It shared homology with bovicin 255 (69%) from Streptococcus bovis and garvicin Q (52%) from Lactococcus garviae. Agilicin C7 has the N-terminal NGY and central YxVTK motifs, a typical feature of Class IId bacteriocins.
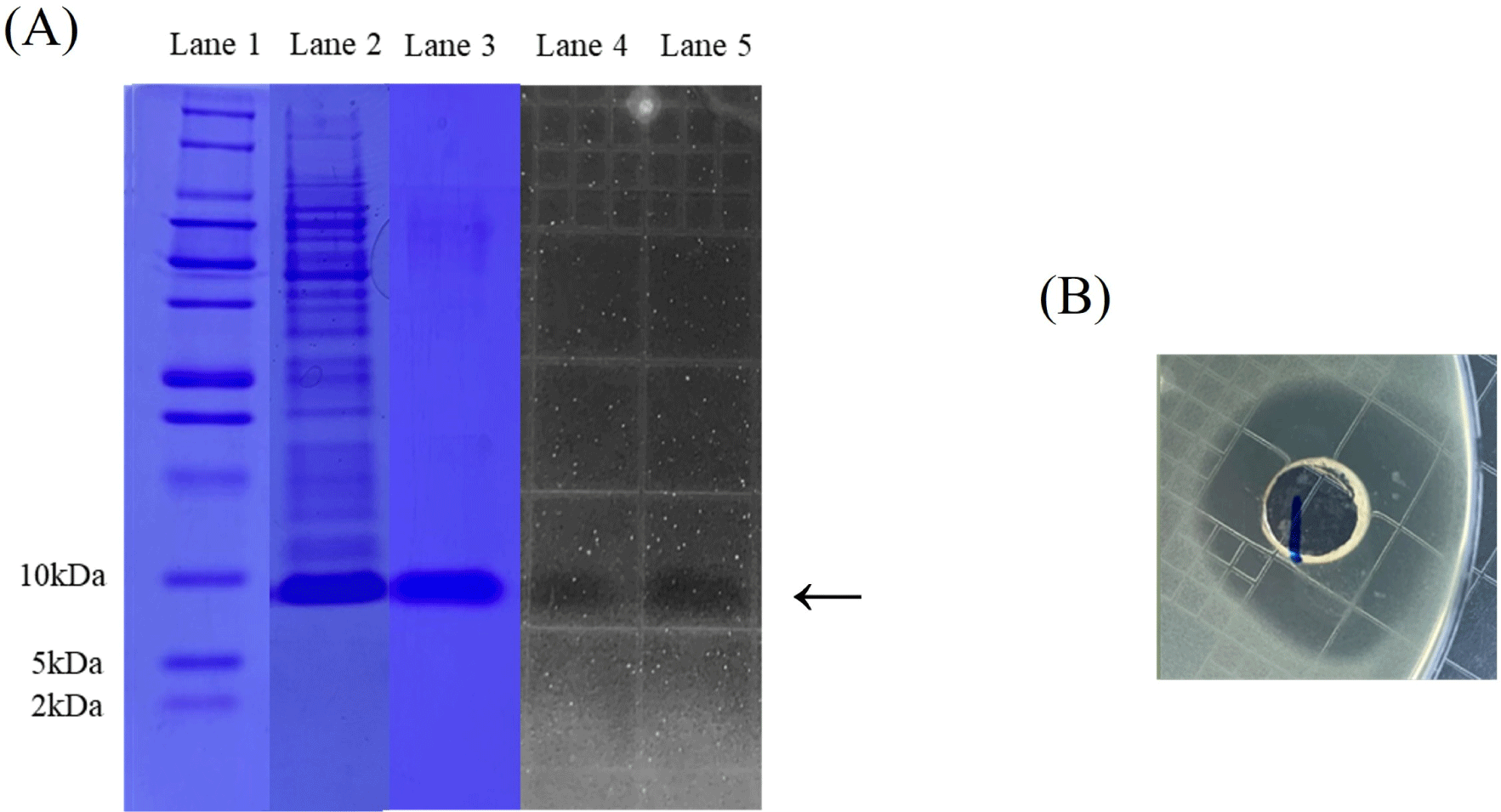
We attempted to express agilicin C7 in E. coli for characterization heterologously. Because sequence analysis of agilicin C7 showed an Ala-Ser-Ala cleavage site at the N-terminus of the signal sequence (Biet et al., 1998; Worobo et al., 1995), mature sequences of the agilicin C7 gene without signal sequences were amplified from the genomic DNA of L. agilis C7 using PCR. The PCR product of agilicin C7 was inserted into the NdeI-SalI site of the pET-21b expression vector, and the resulting plasmid was introduced into E. coli BL21 (DE3) as an expression host by electroporation. Recombinant agilicin C7 was expressed as an inclusion body under IPTG induction and purified using a His-tag purification column (data not shown). Figs. 3A and B shows that r-agilicin C7 exhibits anti-listerial activity. In addition, tricine-SDS-PAGE and zymography (Fig. 3A) showed that approximately 10 kDa of r-agilicin C7 was expressed, confirming its inhibitory activity against L. monocytogenes.
Since r-agilicin C7 inhibited the growth of L. monocytogenes, it was tested whether it showed antimicrobial activity against other bacteria and fungi. As shown in Table 1, r-agilicin C7 inhibited the growth of other listerial species, including Listeria grayi, Listeria innocua, Listeria welshimeri, and Enterococcus spp. However, it did not show antimicrobial activity against the other gram-positive and gram-negative bacteria tested or fungi.
The stability of the r-agilicin C7 against enzymes, heat, pH, and organic solvents was also tested. As shown in Table 2, the anti-listerial activity of r-agilicin C7 was lost by pepsin, α-chymotrypsin, protein K, and trypsin, indicating that agilicin C7 is a proteinaceous substance. Interestingly, its activity lost by α-amylase also lost its activity. These results suggested that agilicin C7 is a glycoprotein. However, SDS-PAGE analysis did not show decrease in the molecular weight of r-agilicin C7 after α-amylase treatment (data not shown), perhaps due to small change in molecular weight. It is necessary to detect the molecular weight change of r-agilicin C7 by α-amylase treatment using liquid chromatography-mass spectrometer in the future, to support that agilicin C7 is glycoprotein.
The anti-listerial activity of r-agilicin C7 was maintained, even after treatment at 100°C for 15 min. In addition, the inhibitory activity was stable in the pH range of 1–10. Furthermore, the anti-listerial activity of r-agilicin C7 was stable after treatment with several organic solvents. These results indicate that agilicin C7 has wide pH and thermal stability, which are features of Class II bacteriocins (Negash and Tsehai, 2020).
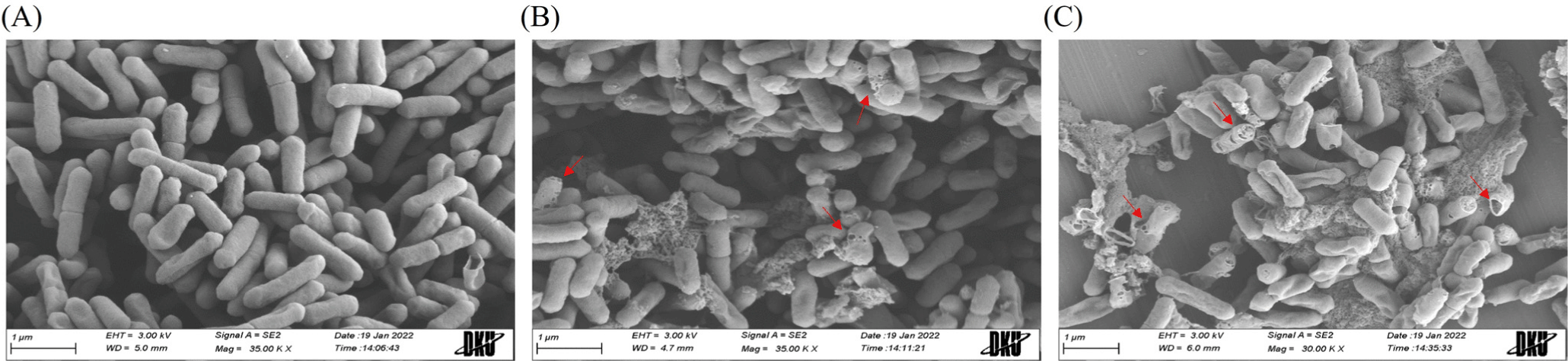
To understand the action mode of r-agilicin C7, FE-SEM analysis was performed to observe cell morphological changes of
L. monocytogenes as an indicator strain. Intact rod-shaped cell structures were observed before r-agilicin C7 treatment (Fig. 4A). The cells appeared extensively wrinkled and porous after treatment with r-agilicin C7 for 2 h, and some cells were lysed entirely, extruding the intracellular material (Figs. 4B and C). Additionally, the degree of cell lysis was dose dependent. Based on these results, r-agilicin C7 destroyed L. monocytogenes by damaging the integrity of the cell envelope, which is a typical property of class II bacteriocins.
Discussion
In this study, we isolated a novel class IId bacteriocin, agilicin C7, from L. agilis. Several L. agilis strains have been reported to exhibit inhibitory activities against foodborne pathogens (Gudiña et al., 2015; Shi et al., 2021). However, information on antimicrobial substances on the biochemical and molecular basis needs to be included. This is the first study to characterize the genetic and molecular properties of an anti-listerial bacteriocin isolated from L. agilis.
Comparative genomic analysis of L. agilis C7 revealed a complete operon for putative bacteriocin biosynthesis, which is highly homologous to the operon for garvicin Q biosynthesis. The agilicin C7 biosynthesis gene contains the N-terminal NGY and central YxVTK motifs, a typical feature of class IId bacteriocins (Tymoszewska et al., 2017; Tymoszewska et al., 2020). Our data show that r-agilicin C7 remained active over a broad pH range and was thermally stable. It was also stable under several organic solvent treatments, all typical properties of class II bacteriocins (Negash and Tsehai, 2020).
Class II bacteriocins have been reported to have bactericidal and pore-forming modes of action (Simons et al., 2020). Class IId is a heterogeneous group of linear, unmodified, non-pediocin-like peptides (Iwatani et al., 2011). Tymoszewska et al. (2017) reported that garvicin Q, a class IId bacteriocin, targets Man-PTS in target cells. The nonpediocin-like class IId bacteriocin, lactococcin A, destroyed target cells through the pore formation mode of action (van Belkum et al., 1991). In this study, L. monocytogenes cells were extensively wrinkled and porous in response to r-agilicin C7. Some cells were lysed entirely, extruding intracellular material, suggesting that agilicin C7 has the same or a similar mode of action. To date, multiple Man-PTS systems have been discovered in L. monocytogenes (Aké et al., 2011), and they control not only sugar transport activity but also the expression of virulence genes (Gaballa et al., 2019). The host specificity of bacteriocins is determined by their outer membrane receptors and inner membrane proteins, which act as scaffolds for bacteriocin insertion and pore formation (Etayash et al., 2016).
Interestingly, the antimicrobial activity of r-agilicin C7 was abrogated by amylase. To date, several amylase-sensitive bacteriocins have been reported (Kang and Kim, 2010; Keppler et al., 1994; Seo et al., 2014), most of which are classified as class IV (Heredia-Castro et al., 2015; Lee and Kim, 2010; Todorov, 2010) or unclassified (Hernández et al., 2005; Seo et al., 2014). Glycosylated bacteriocin is termed “glycocin” (Stepper et al., 2011), which is a cysteine-containing glycoactive peptide in which the O- or S-linked glycan appears essential (Balciunas et al., 2013). However, agilicin C7 does not have these properties, nor does it show sequence homology with glycocin. Therefore, agilicin C7 can be recognized as a novel subgroup of class IId bacteriocins, although the classification of bacteriocins is still controversial. It is necessary to compare the molecular weight change of r-agilicin C7 before and after α-amylase treatment to support that agilicin C7 is glycoprotein, and which amino acid of agilicin C7 is glycosylated in the future.
Many bacteriocins from LAB have been characterized, and these studies have indicated potential applications of bacteriocins in food preservation and safety. Agilicin C7 has physicochemical stability and solid inhibitory activity against L. monocytogenes, suggesting that it can be applied to control L. monocytogenes in the food industry, including dairy and meat products (Yang and Moon, 2021). In the present study, successful cloning and expression of agilicin C7 in E. coli may facilitate mass production of this bacteriocin in high concentration and permit its use as safe antimicrobial food additive. However, whether agilicin C7 can inhibit L. monocytogenes in food matrices need to be investigated in the future because the efficacy and stability of a bacteriocin in foodstuffs are relevant to the chemical and physical properties of foods (Gálvez et al., 2007).













