Introduction
In Europe, starter play a pivotal role in the culinary landscape, especially in the production of dry-cured meats and cheeses. Southern European countries like Italy, Spain, and Southern France are renowned for their extensive use of molds in meat processing, showcasing a tradition that highlights the region’s gastronomic heritage (Spotti et al., 2008). These countries employed the unique properties of molds to enhance the flavor, texture, and preservation of their traditional meat and cheese products, exemplifying the widespread appreciation and application of mold fermentation in enhancing food quality and safety (El-banna et al., 1987; Geisen et al., 1992). For meat fermentation, employing specific mold strains, such as Penicillium nalgiovense for salami and salchichon not only imparts distinctive tastes and aromas but also inhibits the growth of harmful bacteria, ensuring the meat is safe for consumption (Bernáldez et al., 2013; Magistà et al., 2016).
Starters, including bacteria (notably lactic acid bacteria and coagulase-negative staphylococci), yeasts, and molds, are crucial for fermenting meat products, enhancing safety through rapid acidification and antimicrobial substance production. These starters not only help standardize product properties and reduce ripening times but also mitigate microbiological risks from foodborne pathogens (Salmonella spp., Listeria spp., etc) and chemical hazards like biogenic amines, nitrosamines, polycyclic aromatic hydrocarbons, and mycotoxins. Specifically, the lactic acid bacteria reflect their extensive involvement in metabolic activities during fermentation (Radulović et al., 2011). They create an acidic environment by lowering the pH to 4.6–5.9, thereby influencing meat color, and texture, and fostering both fermentation and drying.
The careful selection of non-toxigenic mold strains is essential, as it guarantees the health benefits of fermented meats without compromising safety. Thus, mold fermentation stands as a pivotal practice in the food industry, marrying tradition with modern safety and quality standards (Laranjo et al., 2019). Strains, such as P. nalgiovense and Penicillium chrysogenum, are used to protect dried fermented sausages in the meat processing industry (Bernáldez et al., 2013). Fungal starters have antioxidant effects, prevent spoilage-induced discoloration, enhance texture, generate unique flavors through fat and protein degradation, and influence the appearance of sausages (Philipp and Pedersen, 1988). Moreover, fungal starters are protective against pathogenic and spoilage microorganisms (Geisen et al., 1992; Leistner, 1994; López-Díaz et al., 2001; Singh and Dincho, 1994).
Recently the domestic market for fermented meat products, including fermented sausages, increased in South Korea. Consequently, the demand for fungal starters continues to increase. P. nalgiovense is the only domestically approved fungal starter strain. Importation of foreign strains is necessary for the utilization of starter products in fermented meat products. However, research on fermented sausages has predominantly focused on investigating the impact of additives on sensory quality, sodium alternatives, and storage function (Yim et al., 2020a; Yim et al., 2020b; Yoon et al., 2018). Consequently, the capability and benefits of starter strains derived from local sources remain significantly underexplored (Chung et al., 2017; RDA, 2018). In accordance with the Nagoya Protocol on Access to Generic Resources and the Fair and Equitable Sharing Arising from their Utilization to the Convention on Biological Diversity, the production of fermented foods using domestic bacteria is very important and essential for the fermented food industry. This study aimed to identify domestically derived starter strains suitable for fermented sausages to address the need for homegrown starter development to advance domestic fermented meat production.
Materials and Methods
The fungal starter strains were isolated from traditional fermented foods in South Korea, including Nuruk and Jeotgal, and from mudflats located in Seondo-ri, Hwawangsan, Geumjeong, Soyulgok, Biin-myeon, Seocheon-gun, and Chungcheongnam-do. Isolates were inoculated on potato dextrose agar (PDA) and incubated at 25°C for 7 days. A total of 72 separate strains were isolated. Two isolates from the mudflat were identified as P. nalgiovense and were designated SD01 and SJ02 (P. nalgiovense SD01 and P. nalgiovense SJ02). To prepare spore suspensions, spores were harvested and mixed with 10 mL of 0.1% (w/v) sterile peptone water. Mold600 (M600, Chr. Hansen, Hørsholm, Denmark) was used as the control. All strains were inoculated at a concentration of 8 Log colony-forming units (CFUs). Each strain was stored in 20% glycerol solution at –80°C until required.
For strain identification, lactophenol cotton blue staining at the genus level, internal transcribed spacer (ITS) sequencing, and phylogenetic analysis confirmed P. nalgiovense species. The ITS sequences of the isolates were amplified by polymerase chain reaction (PCR) and analyzed at Macrogen (Seoul, Korea). PCR was performed as described previously (Frisvad et al., 2013). β-tubulin (BenA) and the ITS sequences were generated and combined with sequences used in Table 1. DNA was extracted using a ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, Irvine, CA, USA) as previously described (Ojo-Okunola et al., 2020). PCR was performed using a total volume of 20 μL. The reaction mixture included 200 ng/μL of DNA solution, 10 μL of pre-mixture (AccuPower® Taq PCR Master Mix, Bioneer, Daejeon, Korea), 1 μL of each primer, and RNase-free water. PCR for β-tubulin involved an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, extension at 72°C for 1 min, and a final extension cycle at 72°C for 7 min. The PCR sequences were detected using BLAST in the GenBank database located at the National Center for Biotechnology Information and compared with ITS sequences of previously reported strains. Phylogenetic analysis was performed to determine the genetic relationship of SJ02 within the P. nalgiovense using MEGA software (version 4.0; http://www.megasoftware.net). The neighbor-joining method was used to construct a phylogenetic tree using the maximum composite likelihood model (Kimura, 1980; Saitou and Nei, 1987).
Positive control strains were procured from the Korean Collection for Type Cultures (KCTC, Jeongeup, Korea). They included Penicillium roquefortii Thom (KCTC 6080) and P. chrysogenum (KCTC 6933). Aflatoxin, Ochratoxin A, Patulin, Sterigmatocystin, Cyclopiazonic acid, Penicillin (pcbAB), Penicillin (penDE), Mycophenolic acid, Roqueforine C, and Penicillin (pcbC) β-tubulin were analyzed by PCR using the primers listed in Table 1 (Bernáldez et al., 2013; Färber and Geisen, 1994; López-Díaz et al., 2001; Moavro et al., 2019; Rodríguez et al., 2012). PCR for Aflatoxin (omt-1), Ochratoxin A (otanpsPN), Patulin (idh), Sterigmatocystin (fluG), Cyclopiazonic acid (dmaT), Penicillin (pcbAB), Penicillin (penDE), and Penicillin (pcbC) involved an initial denaturation step at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 1 min, extension at 72°C for 1 min, and a final extension cycle at 72°C for 5 min. For Mycophenolic acid (mpaC) and Roqueforine C (rds/roqA), PCR was performed as just described with annealing temperatures of 54°C and 50°C, respectively. Fluorescence amplification was performed to verify the PCR products.
Extracellular proteolytic and lipolytic enzymes were prepared from the proteolytic broth (Proteo M), as previously described (Biaggio et al., 2016). Proteo M contained (per L) 6.392 g of KH2PO4, 0.522 g of K2HPO4, 0.2 g of MgSO4·7H2O, 1 g of yeast extract, 5 g of NaCl, and 5 g of skim milk powder. In addition, extracellular lipolytic enzymes were prepared from the lipolytic broth (CHO lipo) as previously described. CHO lipo contained (per L) 6.0 g of KH2PO4, 1.0 g of MgSO4·7H2O, 2.0 g of (NH4)2SO4, 4.0 g of peptone, and 10 mL of Tween 20. The medium was adjusted to pH 6.0 and autoclaved for 15 min at 121°C. Extracellular proteolytic and lipolytic enzyme production for each Penicillium species was performed in 50 mL of Proteo M or CHO lipo inoculated with 1 mL (106 spores/mL) of each Penicillium species. They were incubated at 25°C for 14 days with shaking at 150 rpm. After incubation, each production broth was centrifuged at 11,200×g for 5 min. Each supernatant was filtered through a 0.22 μm syringe filter to obtain the crude cell free extract (CCFE), which was assayed. The enzymatic activity of CCFE was assessed on sodium-proteolytic (SP) and lipolytic-tween20 (LT20) plates. A central well was created in the agar plates using a 17 mm cork-borer. CCFE (500 μL) was added to each well. The plate assays were performed in triplicate. The plates were incubated at 25°C overnight and observed for the presence of a clearance halo for proteolytic activity or a precipitation zone for lipolytic activity.
The proteolytic activity was determined as previously described (Magistà et al., 2016). Media were prepared to measure extracellular protease activity, as shown in Table 2. To ascertain the best temperature for maximizing proteolytic activity and growth, the strain was inoculated on skim milk agar plates and maintained at 18°C and 12°C for 9 days. The strain suspension was inoculated at two spots with 5 μL of a suspension containing 104 spores/mL. The enzymatic activity indices for the proteolytic plate assays were determined following the previously described method (Hankin and Anagnostakis, 1975). The semiquantitative enzymatic index (EI) was calculated as EI=H/C, where H represents the diameter of the degradation halo and C is the diameter of the fungal colony. The halo zone around the colony was measured (mm) daily over a span of 9 days, using a ruler along two diametrically opposed directions (Lumi Abe et al., 2015).
The lipolytic activity was determined as previously described (Magistà et al., 2016). To measure the extracellular lipase activity, media were prepared as shown in Table 2. Each strain was inoculated onto plates containing 1% Tween 20 (LT20) or 1% Tween 80 (LT80) and incubated at 18°C and 12°C for 9 days. The strain suspension was inoculated at two spots with 5 μL of a 104/mL spore suspension. The enzymatic activity indices for the lipolytic plate assays were determined as previously described (Hankin and Anagnostakis, 1975). The semiquantitative EI was calculated as EI=H/C, where H represents the diameter of the visible precipitation zone produced by the formation of calcium salt crystals from the enzymatic hydrolysis of the ester bond in the presence of lauric acid (Tween 20) or oleic acid (Tween 80), and C is the diameter of the fungal colony. The precipitation zone around the colony was measured (mm) daily over a span of 9 days using a ruler along two diametrically opposed directions (Lumi Abe et al., 2015).
To determine optimal growth conditions, the strains were inoculated onto meat agar prepared using fermented sausage recipes and NaCl agar prepared by adding different concentrations of NaCl (1%, 2%, 4%, and 6% w/v) to malt extract agar. Each isolate was inoculated with 2 μL (108 spores/mL) at three spots on both the meat and NaCl plates. Subsequently, the plates were incubated at 25°C for 5–7 days and the growth size was determined using Image J software.
To evaluate the fermented sausages manufactured with the identified SJ02 strain, we prepared dry fermented sausages. A mixture of minced pork hindlegs (85%) and fat (15%) was seasoned with salt (2.5%), sodium nitrite (0.015%), pepper (0.3%), red wine (0.4%), dextrose (0.5%), and coriander (0.05%). Additionally, a 0.0125% LAB starter (TRADI 302, Chr. Hansen) was integrated into the mixture. The meat batter was then carefully packed into fibrous casings with a diameter of 40 mm, and the surfaces of the casings were punctured using a sausage pricker. Suspensions of fungal starter cultures (M600 and SJ02) were meticulously adjusted to a concentration of 108 spores/mL and evenly applied to hanging sausages using liquid mold spray (approximately 150 mL). The sausages were incubated for 48 h at 25°C with a RH of approximately 90%. This was followed by a subsequent 4-day drying phase at 18°C and 75% RH. The curing process was approximately 27 days at 12°C and 70% RH. Throughout the manufacturing process, the sausages were continuously monitored for weight loss, which was tracked until they reached 40% (w/w), signifying the successful completion of the fermentation process. Dry fermented sausages were sampled from each batch to evaluate their physical, chemical, microbiological, and sensory characteristics. All assessments were performed three times for each batch.
Detection evaluation of Bacillus cereus, Clostridium perfrigens, Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli O157:H7 was conducted in manufactured sausages. To evaluate microbial safety, 25 g of each sausage sample was aseptically collected, transferred to a stomacher sample bag, and homogenized in 225 mL of the respective suspension media using a model WES-400 Stomacher WiseMix (Daihan Scientific, Wonju, Korea). Appropriate dilutions were spread onto the selective media listed in Table 3 (Kim et al., 2008).
Sausage color was measured in the samples (six 2-cm thick cubic shapes without casing) using a model CR-400 colorimeter (Minolta Camera, Tokyo, Japan) calibrated with the manufacturer’s white calibration plate (CIE L*=93.7, CIE a*=0.3158, and CIE b*=0.3322). The mean values of triplicate measurements for each sample represented the CIE L*, CIE a*, and CIE b* (Essid and Hassouna, 2013).
To test the texture properties, a model TX-700 texture analyzer (Lamy Rheology, Champagne-au-Mont-d'Or, France) was used as previously described (Essid and Hassouna, 2013). More than six cubic samples (2×2×2 cm) of each sample were evaluated. The test was performed by compressing with a cylinder probe (2 cm diameter) at the following conditions: a speed of 1 mm/s, distance of 16 mm, force start of 1 N, wait position at 0.1 mm, up speed of 5 mm/s, and delay for 2 s, while employing the two-bite test methodology, consistent with the sampling technique used for color analysis. The hardness (N), adhesiveness (mJ), cohesiveness (N), gumminess (N), and chewiness (N) of the samples were calculated using texture profile analysis (TPA) curves.
To evaluate the rancidity of the fermented sausages, thiobarbituric acid reactive substance (TBARS) was measured as previously described (Woo et al., 2023). Approximately 3.5 g of the sample was mixed with 500 μL of 0.01% butylated hydroxytoluene and an extraction solvent composed of 7% trichloroacetic acid, 4 N HCl, and 4% HClO4. The homogenate was filtered through the Whatman No. 41 filter paper (Whatman, Little Chalfont, UK). Next, 2 mL of the filtered solution was combined with 2 mL of 20 mM TBA and heated at 80°C for 60 min. After cooling in cold water for 10 min, the absorbance of the solution was measured at 534 nm by using a Multiskan Go spectrophotometer (Multiskan Go, Thermo Fisher Scientific, Waltham, MA, USA). The TBARS values were determined using a standard curve.
Volatile compounds present in the samples were analyzed using the Heracles II electronic nose (E-nose) system (Alpha MOS, Toulouse, France). Approximately 1 g of each sample without casing was placed in a 20 mL headspace vial. The headspace vial were then incubated at 60°C for 20 min while being agitated at 500 rpm to promote the generation of volatile compounds. The collected volatile compounds were coupled to the E-nose system using an automatic sampler. For the gas chromatography analysis, 3 mL of volatile compounds were injected at a speed of 125 μL/s and a temperature of 200°C into the gas injection port connected to the E-nose. The analysis was conducted using MXT-5 and MXT-1701 columns, with an incubation temperature of 50°C and a total analysis time of 10 min. Separated peaks were identified and confirmed using AlphaSoft version 14.2 (Alpha MOS, Toulouse, France) integrated into the E-nose system (Hong et al., 2021).
An Astree electronic tongue (E-tongue) system (Alpha MOS) was used to examine the taste of the samples. The samples were diluted 1:100 in distilled water, homogenized, and filtered prior to analysis. The E-tongue sensor includes five taste component sensors (AHS-sourness, CTS-saltiness, NMS-umami, PKS-sweetness, and ANS-bitterness) and two index sensors (CPS and SCS) that simulate human sensory responses. The sensor values of CPS and SCS were employed for calibration. For analysis, the E-tongue sensor was exposed to the sample extract for 2 min, and the strength of the sensor response to individual taste components was measured through contact. To ensure accuracy and prevent cross-contamination, the individual taste component sensors were washed with purified water for each analysis. The results were verified using AlphaSoft version 14.2 (Hong et al., 2021; Tian et al., 2020).
All experiments were conducted in at least three replicates. Enzyme activity assays, growth assays, color analyses, and sensory evaluations were performed by one-way analysis of variance using the GraphPad Prism 9 software (GraphPad Software, La Jolla, CA, USA). TPA and TBARS assays were performed using independent t-tests. Significant differences between data were determined using Dunnett's tests (p< 0.05).
Results and Discussion
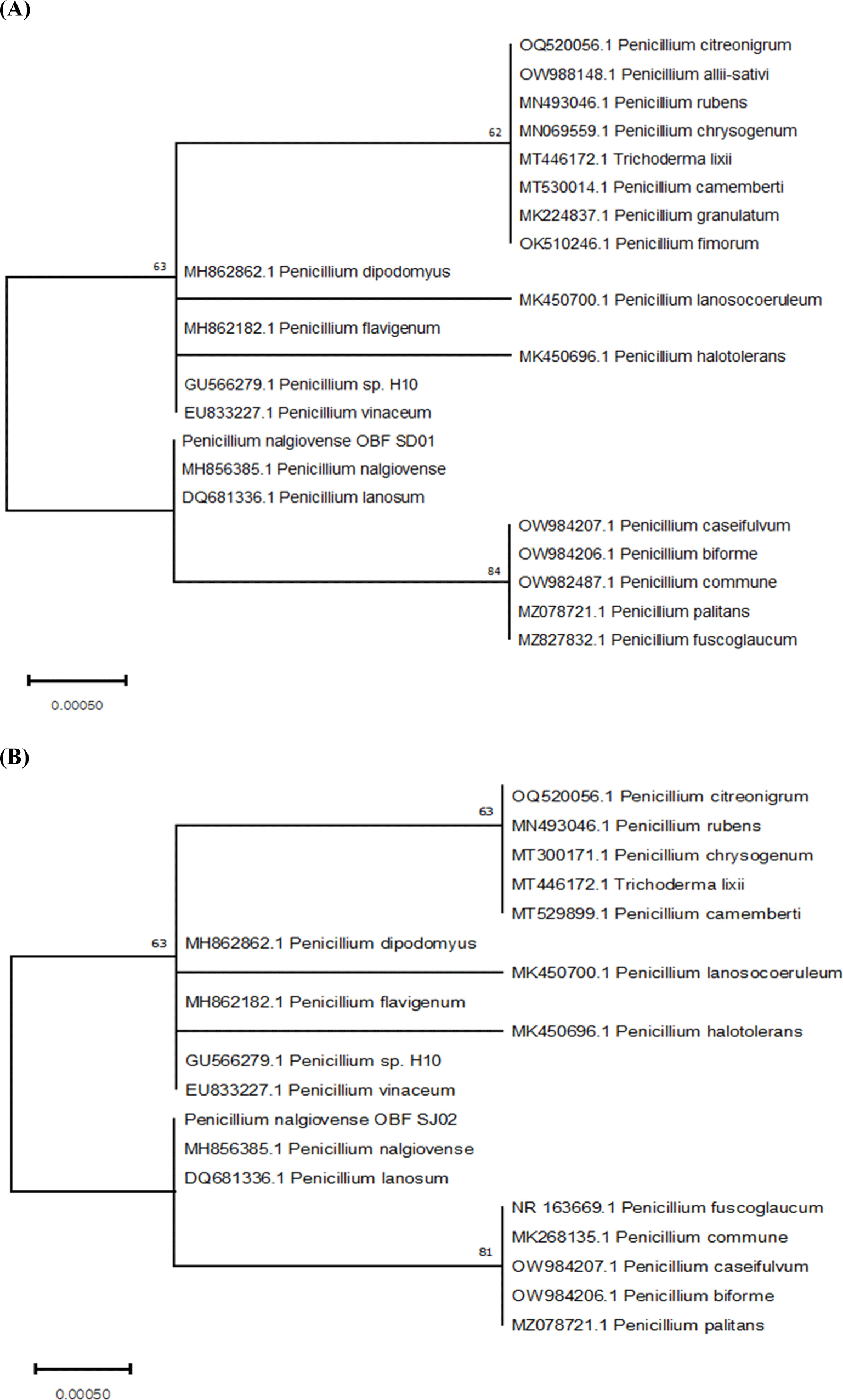
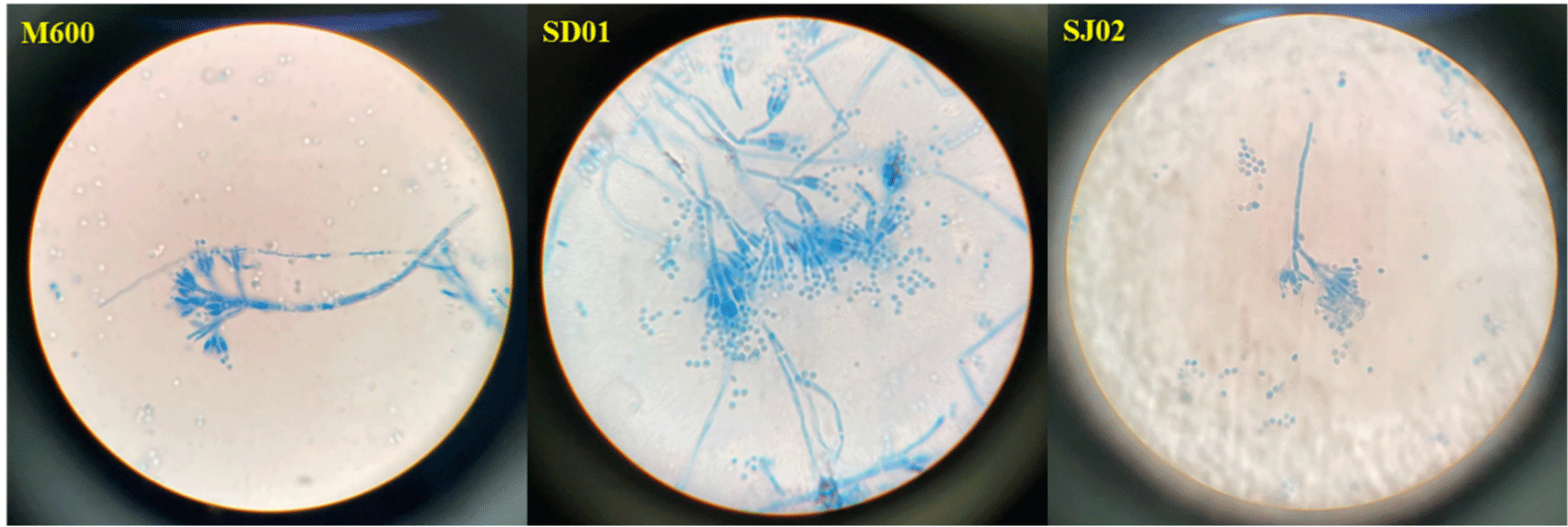
Of the 72 strains obtained from Nuruk, Jeotgal, and the mudflats, 24 were chosen based on their colony morphology. They all belonged to the genus Penicillium (Table 4). The two strains were definitively identified as P. nalgiovense by ITS sequencing. The resulting phylogenetic tree was presented in Fig. 1A for P. nalgiovense OBF SD01 and Fig. 1B for P. nalgiovense OBF SJ02. Microscopic analysis of the conidial morphology of the two selected strains confirmed the characteristic Penicillium conidial shape (Fig. 2). Under the microscope, the combination of septate hyphae, branching at acute angles, erect conidiophores with radiate conidial heads, and chains of conidia give Penicillium its unique and recognizable appearance. These strains were designated P. nalgiovense OBF SD01 (KACC83057BP) and P. nalgiovense OBF SJ02 (KACC83058BP) and were formally deposited in the Korean Agricultural Culture Collection (KACC).
Mycotoxin production by the fungal starter is shown in Fig. 3. Consistent with previous studies, the toxin production capabilities of the isolates were evaluated alongside P. roquefortii Thom (KCTC 6080) and P. chrysogenum (KCTC 6933), which are mycotoxin-producing strains as positive strains (Andersen and Frisvad, 1994; El-banna et al., 1987; Laich et al., 1999; Lopez-Diaz and Flannigan, 1997; López-Díaz et al., 2001; Ludemann et al., 2009; Papagianni et al., 2007).
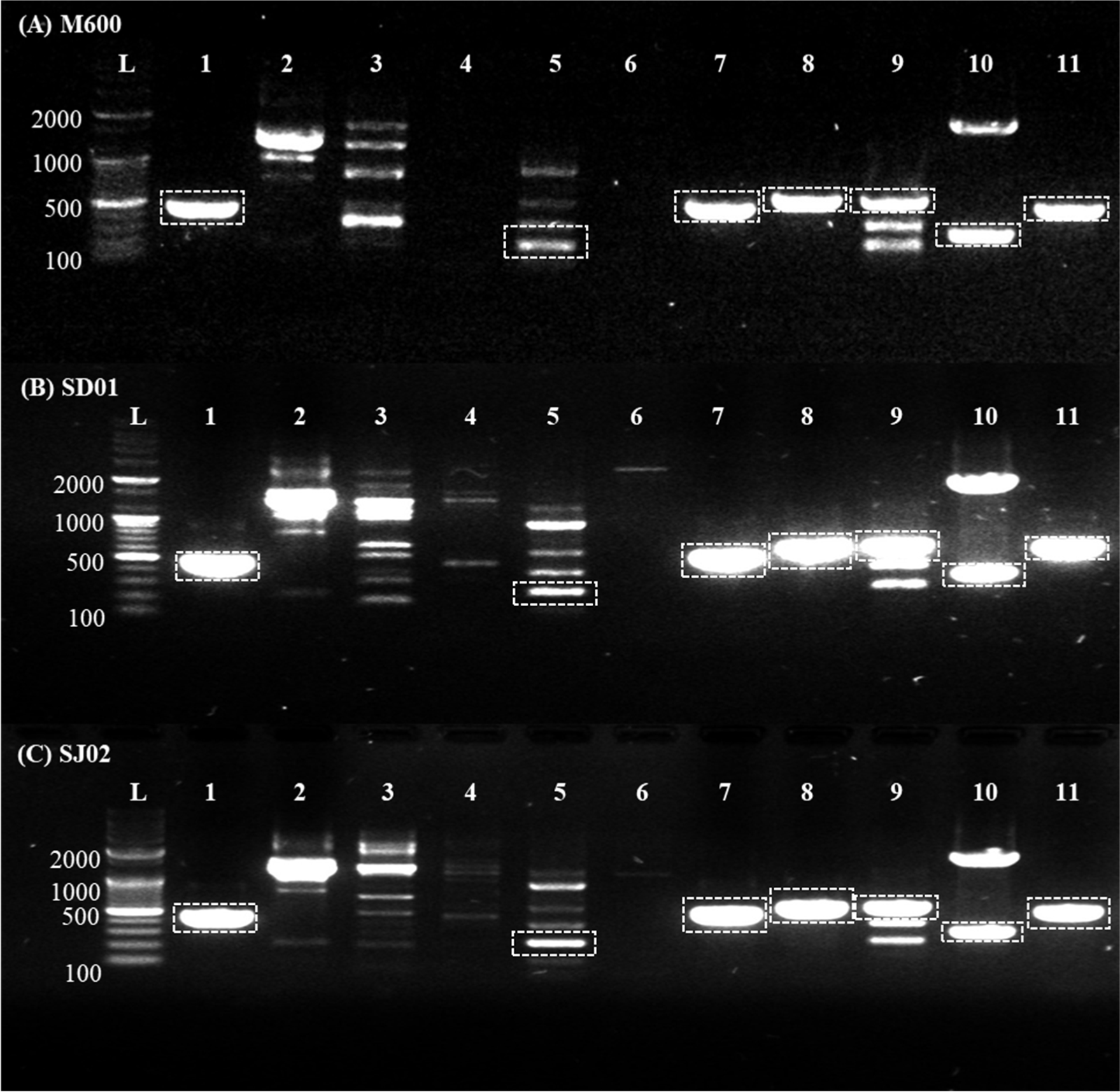
DNA extracted from both P. roquefortii Thom (KCTC 6080) and P. chrysogenum (KCTC 6933) as positive controls and selected strains was identified as belonging to the genus Penicillium. This identification was based on the amplified sequences of the ITS and β-tubulin housekeeping genes specific to P. nalgiovense (data not shown). The expression of two PC strains producing Mycophenolic acid and Penicillin were also confirmed. Upon evaluation of the isolated strains SD01 and SJ02, it was observed that the major mycotoxins producing genes including Aflatoxin, Ochratoxin A, Patulin, and Cyclopiazonic acid, were not detected. However, Sterigmatocystin (fluG), Penicillin (pcbAB), Penicillin (penDE), Mycophenolic acid (mpaC), Roqueforine C (rds/roqA), and Penicillin (pcbC) was detected. Profiles of genes exhibited patterns similar to those of the M600 commercial strain. Despite P. nalgiovense being the most frequently used starter culture for aged and fermented meat products, the fact that this fungus can secrete penicillin into the meat product underscores the importance of obtaining strains that are incapable of synthesizing this antibiotic (Andersen and Frisvad, 1994). It appears that the M600 strain can also secrete antibiotics, suggesting the importance of employing gene disruption techniques to further ensure the safety of our strains for application in the food industry (Laich et al., 2003). However, it has been reported that significant amounts of penicillin were found in the casing and the outer layer of salami meat during the early stages of the curing process, coinciding with fungal colonization, but no penicillin was detected in the cured salami (Laich et al., 1999). Therefore, even though the penicillin synthesis genes exist, the potential for them to be eliminated by penicillinase during the ripening of fermented sausages suggests that our strains might be necessary for faster application in the food industry.
The enzymatic activity of protease and lipase plays a crucial role in the manufacturing of dry fermented sausages, significantly influencing both the flavor and texture of the final product. Therefore, evaluating the enzymatic activity is considered essential for discovering new fungal starters (Magistà et al., 2016). To observe the proteolytic and lipolytic enzyme activities of three P. nalgiovense strains (SD01, SJ02, and M600), spores were cultured on SP and sodium-proteolytic-PDA (SPP) media for measuring proteolytic enzyme activity and LT20 and LT80 media for lipolytic enzyme activity. The clearance halo areas around the colonies were measured from day 2 to day 9. Fig. 4, a displays colony corresponding to days 2, 5, and 10, and quantitative values for the EI were presented (Fig. 4B). SJ02 exhibited the earliest clearance halo on SP media, followed by M600 showing similar activity, while SD01 showed the latest activity on day 9. However, in SPP media supplemented with PDA, which contained rich nutrition to fungal growth, M600 and SD01 showed similar levels of activity, with SJ02 displaying slightly delayed activity. The strain not significantly affected by the nutrient environment in terms of proteolytic activity are suggested to be the control fungi M600 and SJ02. Moreover, lipolytic enzyme activity of SJ02 was shown to be the highest in both LT20 and LT80 media, particularly showing a continuous increase trend in LT20.
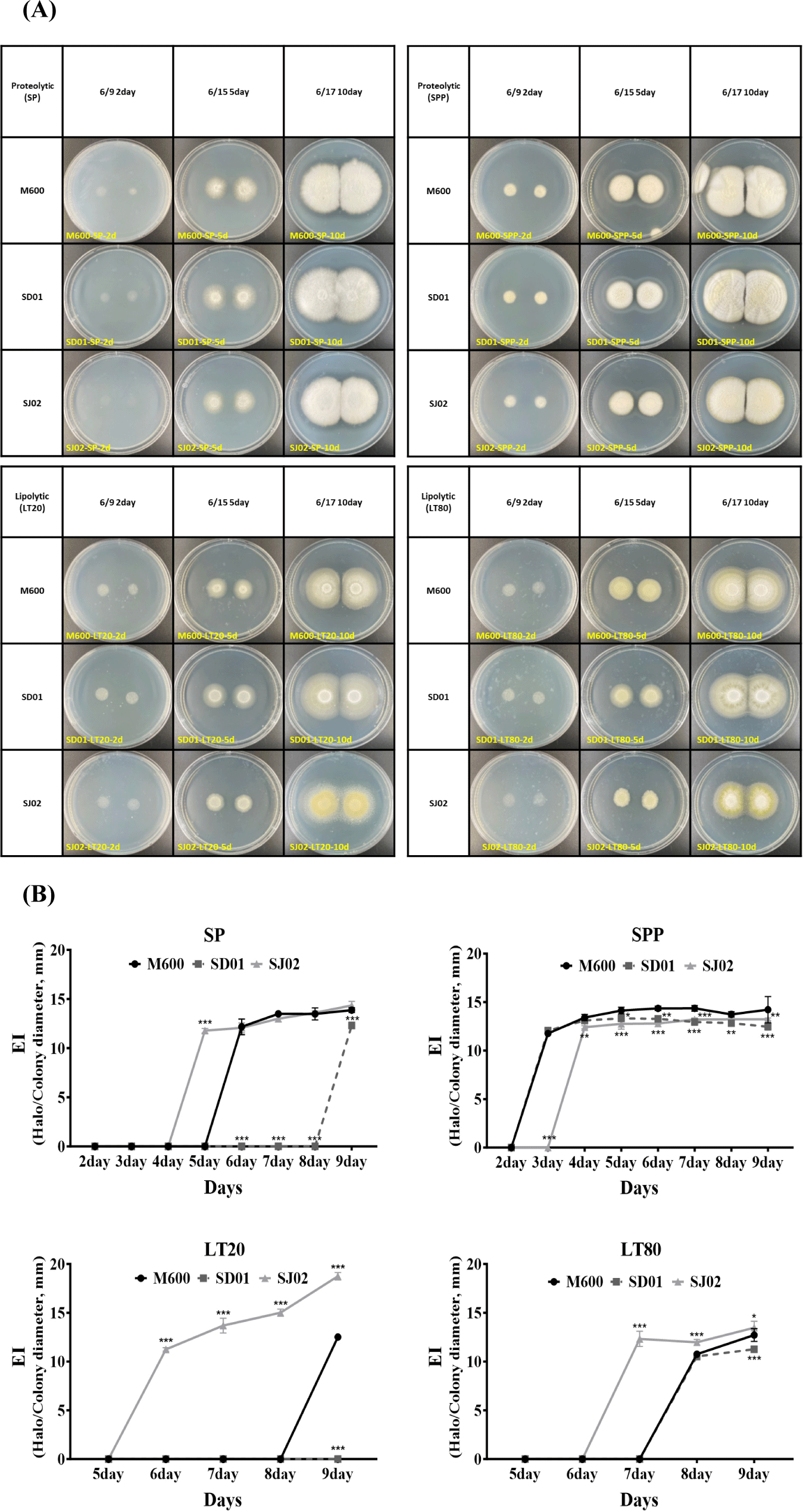
In the CCFE activity assay, the precipitation zone reflects lipolytic enzyme activity. The precipitation zone diameter of SJ02 (2.184 mm) was significantly larger than that of M600 (1.635 mm) at the first day. On the second day, the lipolytic enzyme activity of M600 was 2.168 mm, whereas that of SJ02 was 2.668 mm, confirming that the lipolytic enzyme activity of SJ02 had substantially increased (Fig. 5). These findings suggest that the two isolated strains of P. nalgiovense we identified could potentially contribute more effectively to the breakdown of fat and protein in meat and to the traditional flavor development process than the M600 strain (Toldrá, 2010).
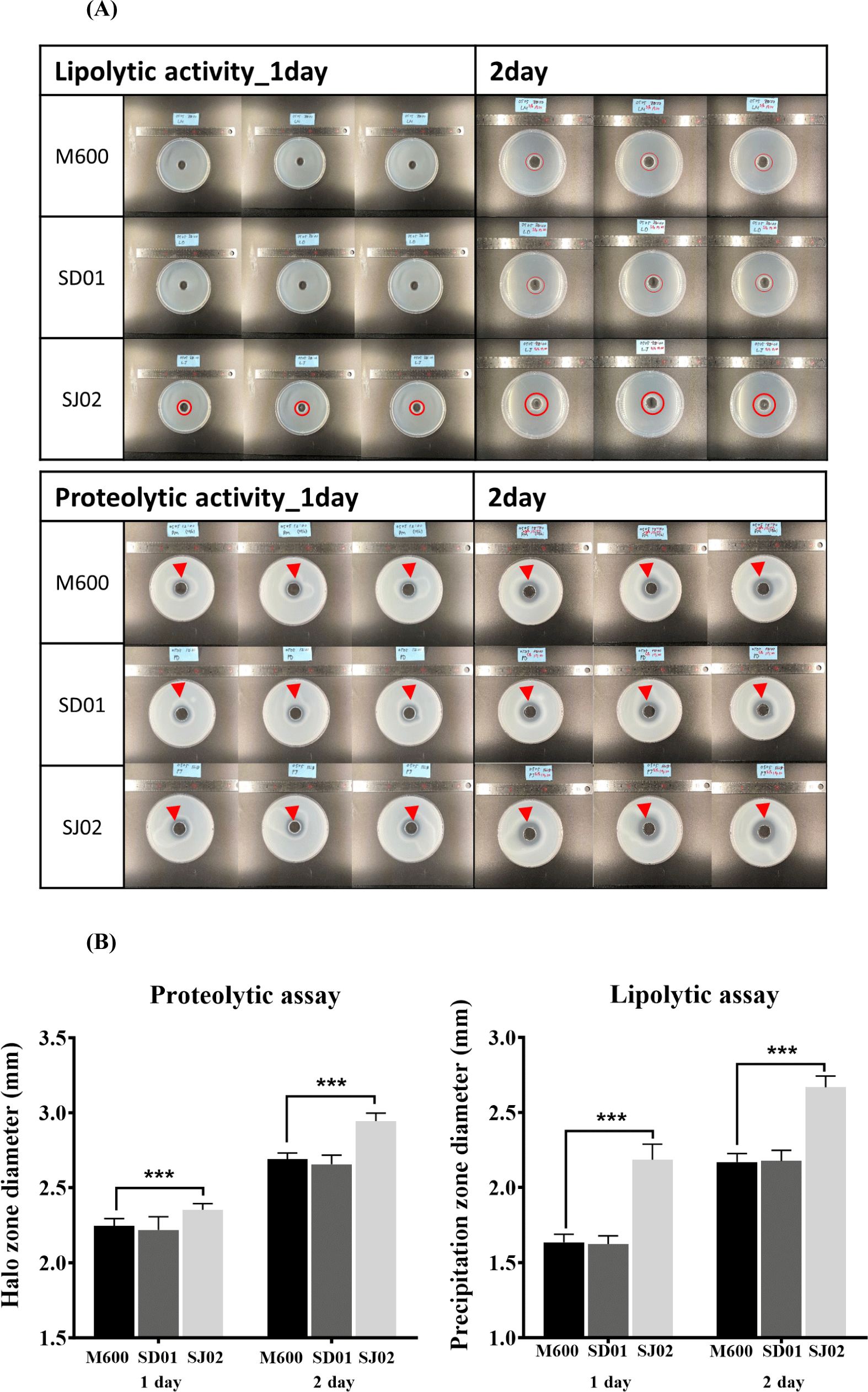
The findings suggest that SJ02, in particular, might be a superior candidate due to its high proteolytic and lipolytic activities, which are crucial for the development of flavor and texture in dry fermented sausages (Galvalisi et al., 2012). The use of strains like SJ02 could contribute to enhancing the quality of meat products by promoting a more efficient breakdown of proteins and fats, thus potentially improving the sensory characteristics of the final product. Recently, there has been active research into utilizing lactic acid bacteria starters, such as Staphylococcus or Lactobacillus, to further enhance the flavor of fermented sausages (Kieliszek et al., 2021; Uppada et al., 2017; Wang et al., 2022). It is necessary to discover Korean fermented food-derived lactic acid bacteria that can provide optimal sensory qualities with SJ02 traits.
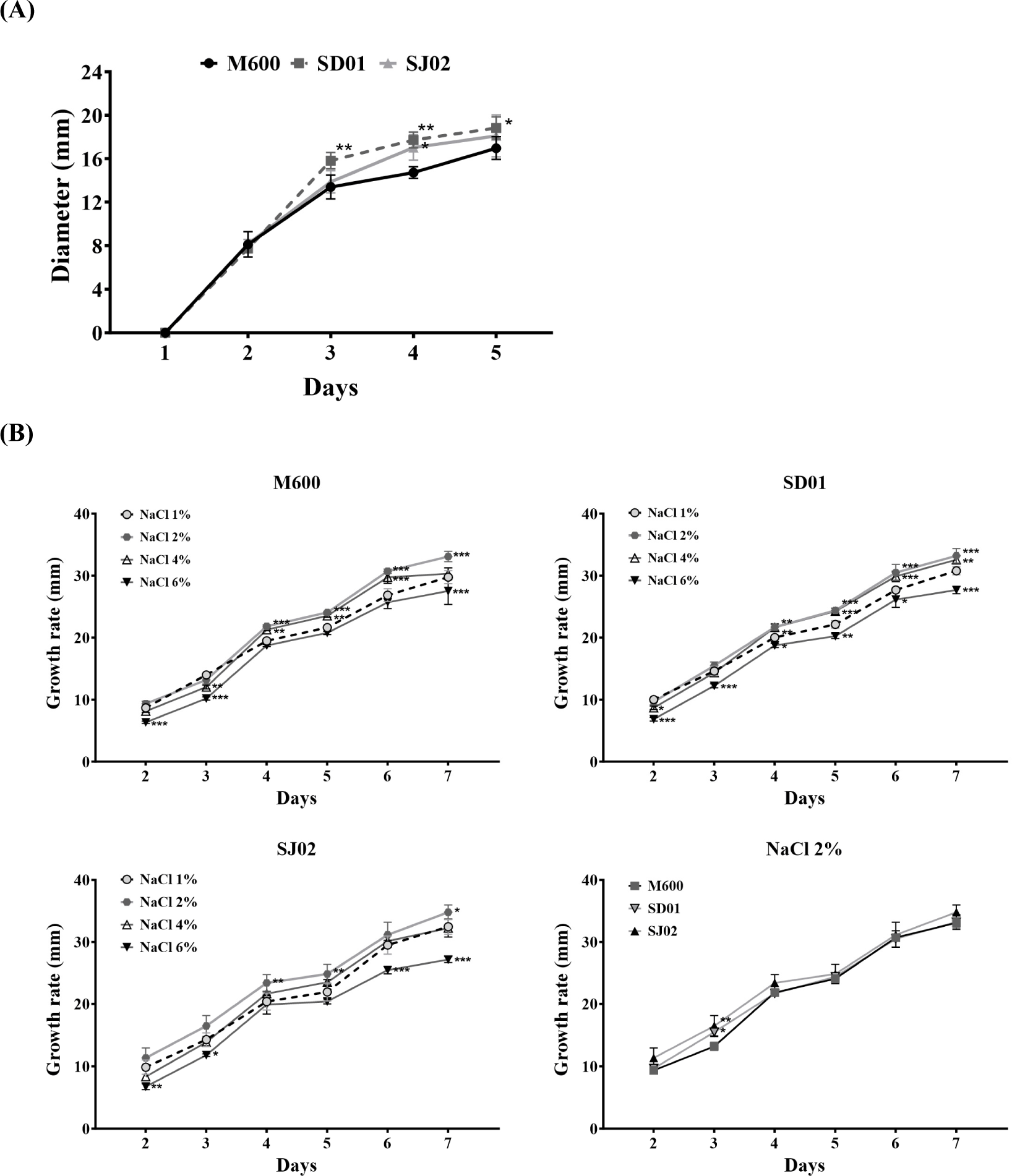
Because P. nalgiovense is widely recognized for its beneficial role in the early colonization of fermented sausages, contributing significantly to the flavor development, prevention of undesirable molds, and appearance and safety of the final product, it is well known that it is very important for candidate fungi to proliferate rapidly in the early stages (Laranjo et al., 2019; Magistà et al., 2016). To compare the growth of fungi without directly manufacturing sausages, we prepared culture dishes under the seasoning conditions as those used for sausage production (Manufacture of dry fermented sausages in Materials and Methods) and inoculated these plates with spores from each strain to observe their growth. The assessment of growth rates on meat plates yielded similar growth patterns for the three strains, establishing strain adaptability and growth performance (Fig. 6). To determine the optimal salinity conditions, the growth rates were compared at different concentrations of NaCl. The growth rates of all three strains were higher at 2% NaCl than at other concentrations. Notably, SJ02 exhibited a significantly higher growth rate (16.47 mm) on day 3 than SD01 (15.48 mm) and M600 (13.19 mm). These results indicate that SJ02 is a promising strain for producing industrially fermented meat products.
To ensure the safety against undesirable bacteria, specifically pathogenic bacteria, when making fermented sausages using SJ02 and M600, both quantitative and qualitative analyses of Bacillus cereus, Clostridium perfringens, Staphylococcus aureus, Listeria monocytogenes, Escherichia coli O157:H7, and Salmonella spp. were examined. Additionally, the mycotoxins Aflatoxin and Ochratoxin A were also analyzed. The microbiological safety and toxin safety of fermented sausages using the SJ02 strain were verified to be as safe as those with the M600 strain. No quantitative changes in the lactic acid bacteria starter named Bactoferm TRADI 302, which contains Lactobacillus sakei, Staphylococcus carnosus, and Staphylococcus xylosus, were observed (Table 5). The application of SJ02 in dry fermented sausage production can ensure food safety by preventing the proliferation of harmful foodborne pathogens (Bernáldez et al., 2017; Büchter et al., 2020; Hankin and Anagnostakis, 1975).
As fatty acid oxidation progresses, levels of malondialdehyde (MDA) and acetal compounds increase. These compounds are correlated with food deterioration and thus serve as parameters of meat product freshness. The reaction of 2-TBA with these compounds produces a colored complex, which is a widely accepted evaluating method of the extent of fatty acid degradation and oxidation. Unsaturated fatty acids are susceptible to oxidation, generating peroxides such as hydroperoxides that can damage proteins and DNA, induce mutations and carcinogenesis, and compromise food quality. A TBARS value exceeding 1 MDA ppm (μg/mL) is considered high rancidity and renders the product inedible (RDA, 2009). The TBARS value for dry fermented sausages with SJ02 was <1 MDA ppm (μg/mL); it is considered safe for fatty acid deterioration in meat products (Table 6).
The dry fermented sausages made with SJ02, identified from domestic source in this study, have consequently similar color and texture characteristics compared with those made with the commercial starter M600. Specifically, no significant differences were observed between M600 and SJ02 sausages. In addition, textural indicators, such as hardness, cohesiveness, gumminess, and chewiness, were not significantly different between sausages made with M600 and SJ02 when analyzed using a texture analyzer. Only adhesiveness value in SJ02 sausages was higher (p<0.05) than that in M600 sausages (Table 6). Notably, one of the main role of fungal starters has been recognized as influencing the flavor and texture of dry fermented sausages (Casaburi et al., 2008). These factors significantly shape consumer preferences and have been actively investigated to enhance the quality of dry fermented sausages (Bourdichon et al., 2012; Fadda et al., 2010). Similar quality characteristics to the current research can be observed in previous studies (Chinaglia et al., 2014; Fadda et al., 2001; Seong et al., 2008; Tian et al., 2020; Witte et al., 1970; Yang et al., 2018; Zhao et al., 2011). It implies that SJ02 found in this study could be as good a fungal starter culture as commercial stater cultures without compromising quality.
The E-nose technology have been used to determine aroma and taste profiles in targeted food products, serving as an indicator of the olfactory and gustatory perceptions experienced by consumers during the consumption of food. The present study used the E-nose technique to systemically evaluate the distinct flavor and taste of dry fermented sausages influenced by each strain (Jeong et al., 2022).
In the E-nose analysis, M600 sample exhibited elevated levels of ethanol and propanal, which are associated with pungent odors, in comparison to SJ02 sample. Conversely, the concentrations of propan-2-one and butyl butanoate, which represent fruity characteristics, were higher in SJ02 than in M600 (Fig. 7). Notably, E-nose analysis revealed that the different utilization of the fungal starters, M600 and SJ02, in fermented dry sausages led to variation in the composition of the final products. These observations suggest the potential for future investigations to identify difference in flavor and taste between sausages manufactured using M600 and SJ02 strains, thus confirming the distinct characteristics of SJ02-isolated fermented sausages in South Korea.
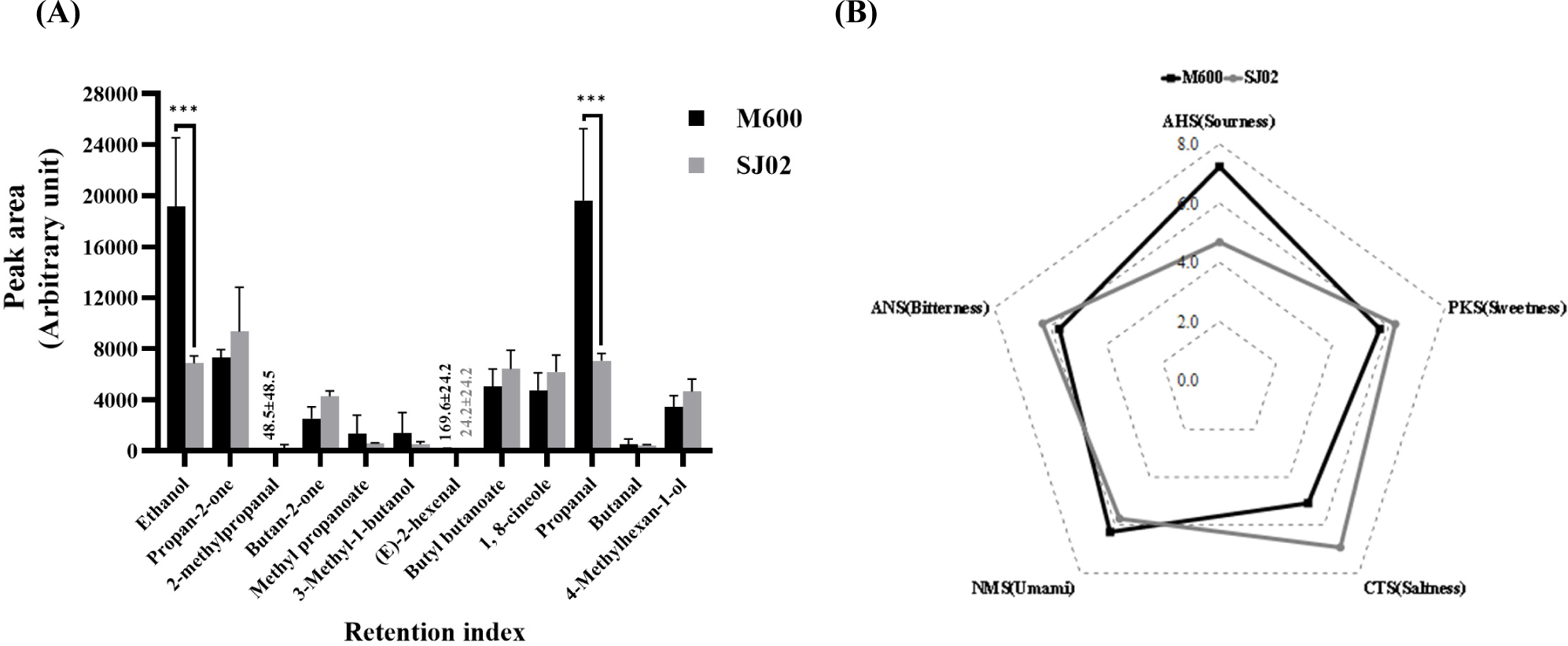
The results of the E-tongue analysis are presented as a radar graph, representing taste profiles measured by the E-tongue in dry fermented sausages using the fungal starters M600 and SJ02 (Fig. 7). Each taste parameter, along with representative taste qualities, was determined using five taste sensors (AHS, PKS, CTS, NMS, and ANS; Ju et al., 2016).
The AHS sensor, which measures acidity and bitterness, primarily captures the acidity in fermented dried sausages. The AHS values were recorded as 7.3±0.2 for M600 and 4.7±2.0 for SJ02, indicating a lower level of bitterness in SJ02 and a higher level of acidity in M600. The CTS sensor, which evaluates saltiness, yielded values of 5.1±0.0 for M600 and 7.0±0.9 for SJ02, highlighting a higher level of saltiness in the SJ02 sample. With regard to NMS sensor responsible for savory and salty tastes, M600 recorded 6.3±0.0, while SJ02 displayed 5.8±0.2, indicating a greater savory taste in M600. Finally, the ANS sensor, which detects bitterness, recorded 5.7±0.2 for M600 and 6.3±0.2 for SJ02, signifying elevated bitterness in SJ02.
When comparing the taste characteristics of fermented dried sausages produced using strains M600 and SJ02 through E-tongue analysis, no statistically significant differences were observed in the measured values between the two strains. The findings imply that fermented dried sausages crafted using the two strains had similar taste characteristics. Therefore, the potential utilization of the SJ02 strain for adjusting the taste attributes of fermented dried sausages in accordance with the microbial strain is suggested. However, further studies are necessary including sensory evaluations conducted by trained panels that consider a range of diverse taste perceptions (Van Ba et al., 2018).
Conclusion
This study identified two P. nalgiovense strains, SD01 and SJ02, as potential starter cultures for fermented sausages from Korean sources. SJ02, in particular, showed enhanced enzymatic activities and growth in high-salinity conditions compared to a commercial starter, M600. Fermented sausages applied with SJ02 exhibited improved enzymatic activity and safety, and we found that SJ02 fermented sausages exhibited similar qualities to M600 fermented sausages, confirming the potential of our isolate SJ02 as a domestic commercial strain to replace M600.













