Introduction
The species Salmonella enterica comprises more than 2,500 serovars, and causes one of the most frequently-reported foodborne disease, salmonellosis. S. enterica infects numerous animal hosts, and is considered as a significant public health threat worldwide. The severity of salmonellosis ranges from mild gastroenteritis to life-threatening typhoid, and depends on the serovar type and the species of infected host animal. Although many nontyphoidal serovars are only associated with relatively minor symptoms, they are still responsible for the largest burden of food borne disease in the world (Crump et al., 2015; Kirk et al., 2015). In addition, S. enterica infects a wide range of host mammals, causing significant zoonoses, which is particularly damaging to the livestock and meat processing industries (L Plym and Wierup, 2006). To control salmonellosis, a variety of antibiotics and chemicals have been widely used in veterinary medicine and in interventions for food processing. However, the emergence of multidrug-resistant S. enterica strains has increased levels of drug tolerance, and necessitates the development of new antimicrobials (Doyle, 2015; Wales et al., 2010). Many antimicrobial drugs or chemical preservatives target the Salmonella cell envelope, the disruption of which efficiently abolishes numerous essential functions. Despite the availability of these drugs, analytical methods validating whether a given chemical can cause damage to the Salmonella envelope are still lacking (Wolf and Mascher, 2016).
In this study, we examined whether expression of the S. enterica serovar typhimurium spy gene could be used as a biosensor for measuring envelope stress. Spy was originally reported to be exclusively expressed in the periplasmic space of Escherichia coli during spheroplast formation, and was therefore named spheroplast protein y (Hagenmaier et al., 1997). Further studies with E. coli revealed that Spy functions as a periplasmic chaperone with high flexibility and its expression is induced when cells are exposed to envelope stresses that result in protein misfolding, such as ethanol, indole, copper, zinc, and spheroplasting (Bury-Mone et al., 2009; Quan et al., 2011; Raffa and Raivio, 2002; Wang and Fierke, 2013). Indole treatment also increases spy transcription in S. typhimurium (Appia-Ayme et al., 2012). These studies strongly suggest that Spy is involved in maintaining protein homeostasis in the periplasmic space in response to various envelope stress conditions, and that measuring the induction of spy transcription may be useful for monitoring envelope stress in bacteria.
In this study, we investigated spy transcription in S. Typhimurium under several envelope stresses, and examined the effect of regulators for three major envelope stress signal transduction systems, RpoE, Cpx, and Bae, on spy transcription.
Materials and Methods
Wild-type (WT) parental strain, S. typhimurium 14028S, and its isogenic gene mutant strains were used. Luria–Bertani (LB) broth (Difco, USA) was used for bacterial culture. For broth culture, strains were cultured in LB broth at 37°C with shaking. To construct gene knockout mutants, the polymerase chain reaction (PCR)-mediated one-step gene mutation using the λ Red recombinase was used, as described previously (Datsenko and Wanner, 2000). baeR and cpxR deletion mutant strains were constructed using the following primer pairs: (baeR) F: 5’-aacacgccgc gcattttgat tgttgaagat gaacccaagc gtgtaggctg gagctgcttc-3’ and R: 5’-caccccgtag accgcgcgta taaatgactg ttcggcatcc catatgaata tcctccttag-3’; and (cpxR) F: 5’-gttagttgat gatgaccgag agctgacttc cctgttaaaa catatgaata tcctccttag-3’ and R: 5’-aaaccacggg tgaccgtctt tgcgttccgg cagtttgcgggtgtaggctg gagctgcttc-3’. Primer pairs, F: 5’-cgtcaatatt gtacacgcgc-3’ and R: 5’-acagaaacgt catctcacc-3’ (baeR), and F: 5’-gatgtgttgc cgtcaacgta-3’ and R: 5’-actggctgga gctcttact-3’ (cpxR) were used for PCR confirmation. Final selection of mutants was performed as described previously (Park et al., 2015).
To construct spy transcriptional fusion, we employed a promoter probe vector plasmid pFPV25, which contains a GFP reporter gene (gfpmut3) reporter gene (gift from Raphael Valdivia) (Valdivia and Falkow, 1996). A DNA fragment containing the spy promoter region was amplified from S. typhimurium chromosomal DNA by PCR using Pfu polymerase (iNtRON Biotech., Korea) with primer pairs F: 5’-tatgagctct gctatatcat gctgttgta-3’ and R: 5’-tatggatcca ggacggctat agaattctctg-3’. The purified PCR products were digested with SacI and BamHI (NEB, USA), and then ligated with SacI/BamHI-digested pFPV25. A single clone was finally selected following DNA sequence confirmation of insert region in pFPV25. S. typhimurium harboring the spy-gfp fusion plasmid was cultured over-night and then diluted 1:1000 in fresh LB medium. The culture was grown to log phase (OD600~ 0.4), and then cells were treated with 4% ethanol or polymyxin B (1 ug/mL) for 1 h. After washing with phosphate buffered saline, the fluorescence intensity of bacterial cells (adjusted to OD600 = 1.0) in a black 96 well plate (SPL, Korea) was measured with excitation at 485 nm and emission at 535 nm using a DTX 880 microplate reader (Beckman Coulter, USA).
Method used for spheroplast formation was modified from a previously described strategy (Birdsell and Cota-Robles, 1967). Briefly, overnight cultures of WT S. typhimurium were diluted 1:4 into fresh LB broth and cultured to mid-log phase (OD600~ 0.8). Cells were collected by centrifugation (5,000 rpm for 20 min at room temperature) were washed once with 10 mM Tris buffer (pH 8.0). Following centrifugation as above, the cell pellet was resuspended with 10 mM Tris buffer (pH 8.0) containing sucrose (0.5 M) and incubated for 10 min before the addition of lysozyme (200 μg/mL). The cells were incubated for 10 min, and then an equal volume of Tris buffer containing EDTA (20 mM) was added and the cells were incubated at 37°C for a further 4 h.
Salmonella cells harboring spy-gfp fusion for microscopic observation were washed once with 10 mM Tris buffer (pH 8.0) before analysis. Cell aliquots were dropped onto microscope slides and coated with 1% agarose, and then analyzed using a Zeiss Axio A1 microscope (Zeiss, Germany) with an oil-immersion 100X objective lens. Images were captured with a charge coupled device camera.
Results and Discussion
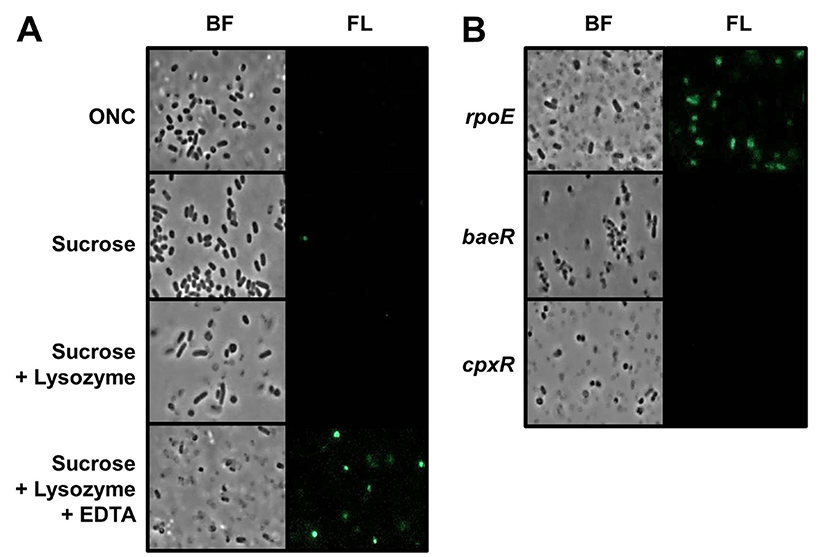
To examine changes in spy transcription in S. Typhimurium during spheroplast formation, we constructed a spy-gfp transcriptional fusion plasmid. This type of operon fusion can be useful for monitoring Spy expression because GFP protein expressed by the spy promoter remains in cytoplasm of spheroplast cells. Natural Spy protein is exclusively expressed in periplasmic space, without any membrane association, and can diffuse out of the cell when outer membrane of spheroplasts were osmotically lysed during spheroplasting (Birdsell and Cota-Robles, 1967; Hagenmaier et al., 1997). To determine whether spy transcription was induced during spheroplast formation in S. typhimurium, the morphology and fluorescence intensity of spy-gfp containing cells were examined by fluorescence microscopy. Fig. 1(A) clearly shows that WT S. Typhimurium spheroplasts were strongly fluorescent. The method used for spheroplast formation in this study essentially involves sequential exposure to high osmotic pressure induced by sucrose, peptidoglycan lysis by lysozyme, and then membrane weakening by EDTA. Of note, cells with rounded morphology were conspicuously fluorescence after all processes for spheroplast formation were completed. Given the fact that removal of cell wall can create rounded spheroplasts, spy transcription in S. typhimurium is likely triggered by changes in membrane rigidity. While E. coli can form spheroplasts without the addition of EDTA (Birdsell and Cota-Robles, 1967), we did not observe rounded spheroplasts and spy-gfp expression in the absence of EDTA. Although it is not known whether induction of spy transcription in E. coli spheroplasts requires EDTA treatment in addition to sucrose and lysozyme, our result suggests that there may be differences in the envelope architecture of two species.
Three typical envelope stress response systems have been identified in E. coli and Salmonella species (Rowley et al., 2006). The alternative sigma factor, RpoE, directs transcription of many genes involved in envelope homeostasis, and two other envelope response systems, Bae and Cpx, both comprising a membrane histidine kinase and a cognate response regulator protein, also promote transcription of specific sets of genes in response to envelope stress. To examine the roles of these systems in spy transcription, we introduced spy-gfp fusion plasmid into S. typhimurium mutants lacking rpoE, baeR, or cpxR. Fig. 1B shows effects of these mutations on spy transcription during spheroplasting. More spheroplast cells were fluorescent in rpoE mutant than in WT, whereas no fluorescent cells were detected in baeR and cpxR mutant strains. These results demonstrate that the induction of spy transcription in S. typhimurium spheroplasts requires Bae and Cpx systems, and that elimination of RpoE-derived envelope protection can increase the spheroplast population, inducing spy transcription.
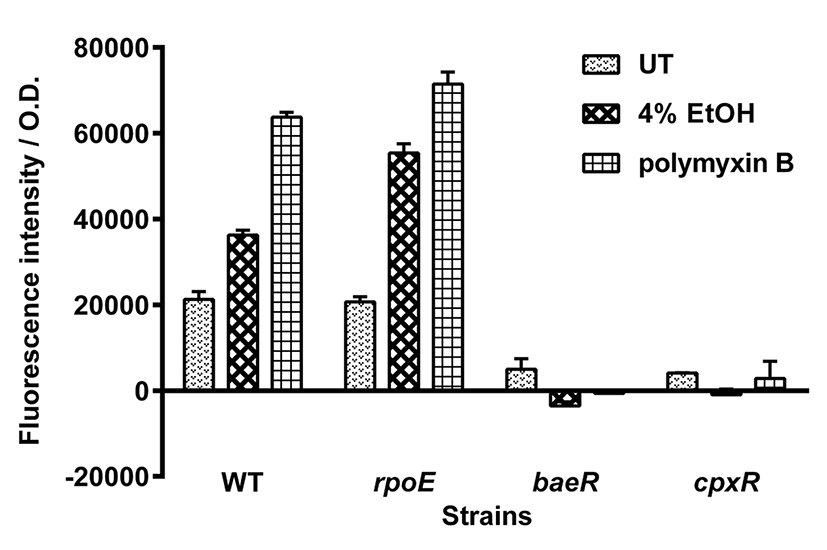
In E. coli, spy transcription is induced by exposure to membrane-disrupting agents, including indole and ethanol (Raffa and Raivio, 2002; Srivastava et al., 2014). To examine whether spy transcription in S. typhimurium is also induced under membrane stress conditions, spy-gfp fusion-containing Salmonella were exposed to 4% ethanol or the membrane-damaging antibiotic polymyxin B. As shown in Fig. 2, S. typhimurium treated with 4% ethanol or polymyxin B showed an approximately 2- or 3-fold increase in spy transcription, respectively, compared with that of untreated cells. In microscopic analysis, the ethanol- or polymyxin B- treated cells mostly showed strong fluorescence, while untreated cells did not (data not shown). As was observed for spheroplasting, spy transcription was enhanced in rpoE mutant under both envelope stress conditions, confirming that spy transcription can be increased in the absence of the envelope protection systems. Again, mutations in baeR and cpxR abolished the induction of spy transcription in ethanol- and polymyxin B- treated S. typhimurium. As was observed in E. coli (Srivastava et al., 2014), spy transcription in S. typhimurium spheroplasts and in cells exposed to 4% ethanol required both the BaeR and CpxR regulators. Both regulators were also required for induction of spy transcription in polymyxin B-treated cells. Thus, regulatory elements for spy gene transcription appear to be conserved between the two species.
In conclusion, this study has clearly demonstrated for the first time that spy transcription can be induced in S. typhimurium by exposure to envelope stresses, including ethanol, polymyxin B, and spheroplasting, and is dependent on the envelope stress response regulators BaeR and CpxR. Homology analysis showed that the spy genes are conserved amongst all serovars of S. enterica, including pathogens causing zoonosis (data not shown). Spy, which functions as a broad-spectrum chaperone, is exclusively expressed in the periplasmic space under envelope stress conditions, which are sensed by two major envelope response systems. Therefore, it is plausible that Salmonella strains harboring the spy-gfp fusion constructed in this study could be used as a whole-cell biosensor for screening the efficacy of new antimicrobials that specifically target the Salmonella cell envelope.













