Introduction
Kocuria salsicia is a coccoid, gram-positive, and facultative anaerobic bacterium (Savini et al., 2010). It has been isolated from various animal hosts, soil, dairy products, the skin or oropharynx mucosa of humans, and high-salt and high-temperature environments (Basaglia et al., 2002). Kocuria spp., such as K. kristinae, K. varians, K. rhizophila, K. rosea, and K. marina, have been demonstrated to cause infections in humans (Lee et al., 2009). Sepsis and increased platelet and leukocyte counts are signs of Kocuria spp. infection (Dunn et al., 2011). Although few studies have evaluated the mechanisms of the infections and toxicity of Kocuria spp., biofilm has been suggested to be involved for adhesion and colonization (Meletis et al., 2012). Infections associated with Kocuria spp. include urinary tract infections, cholecystitis, and catheter-related bacteremia (Kandi et al., 2016). Among Kocuria spp., K. salsicia is the causative agent of the first case of catheter-related bacteremia in Korea (Sohn et al., 2015).
The bacterial growth curve has been demonstrated to be useful in identifying the physiological characteristics of microorganisms; it indicates the phases of bacterial growth and tolerance in certain environments, such as various temperature and salt conditions (Zwietering et al., 1990). Biofilms are formed when microorganisms grow at a particular spot and reach a specific density, which can be predicted via a growth curve. Therefore, it is important to evaluate the growth curve for determining the biofilm-formation ability of the bacteria (Welch et al., 2012). Microbial attachment and biofilm formation on food contact surfaces in processing plants are major concerns in terms of survival of these microorganisms under extreme conditions, such as high osmotic pressure, and the risk of cross-contamination (Ryu and Beuchat, 2005). Recently, Kocuria spp. have gained prominence owing to a rise in the number of reports of human infections, signifying their pathogenic potential (Kandi et al., 2016). The infections caused by Kocuria spp. include urinary tract infections, catheter-associated bacteremia, and endocarditis, which might be associated with their biofilm-forming ability (Moreira et al., 2015; Sohn et al., 2015).
Currently, in farmstead manufacturing, which involves direct handling of livestock, antimicrobials are used to reduce biofilm-forming bacterial contamination; moreover, antibiotic resistance has increased over the past few decades (Mehli et al., 2017). The use of antimicrobials in clinical settings and food production is inefficient; nevertheless, the overuse of antimicrobials has resulted in the development of antimicrobial resistance in bacteria present in livestock products (European Food Safety Authority, 2019). Particularly, microbial contamination during cheese-making may act as an intermediate for transferring antimicrobial resistance genes to various bacteria, including non-pathogenic bacteria, and may lead to multidrug resistance development (Locatelli et al., 2016). Furthermore, these multidrug-resistant (MDR) bacteria act as a reservoir of antimicrobial resistance genes, facilitating their transmission to humans via food (Golob et al., 2019).
Many studies have focused on foodborne pathogens present in raw milk, cheese products, and environments in which farmstead cheese is produced (D’Amico and Donnelly, 2008; Fox et al., 2011; Kang et al., 2018; Mehli et al., 2017). However, no studies have focused on the microbial contamination of cheese brine, possibly because of its high salt concentration. To the best of our knowledge, this study is the first to report the isolation of K. salsicia from cheese brine from a farmstead cheese-manufacturing plant in Korea. The aims of the present study were to evaluate the (i) growth curve of K. salsicia strains isolated from cheese brine at different temperatures (5°C, 15°C, 25°C, 30°C, 37°C, and 42°C) and 15% salt concentration, (ii) biofilm-forming ability of K. salsicia strains at the different temperatures and media [phosphate buffered saline (PBS), nutrient broth (NB), and NB containing 15% sodium chloride (NaCl)], and (iii) antimicrobial susceptibility of K. salsicia strains as well as assess the potential risk posed by farmstead cheese (contaminated by organisms in the brine) with respect to the transfer of antimicrobial-resistant pathogens to humans.
Materials and Methods
From June to December, every month in 2020, a sterile bottle was used to collect two 500 mL bottles of string cheese brine per month (totaling 14 bottles) during cheese-making at a farmstead cheese house located in the Gyeong-gi province, Korea. Salt concentration and pH of the brine were analyzed using a glass salimeter (Daedong, Seoul, Korea) and Orion Star™ A211 pH Benchtop Meter (Thermo Fisher Scientific, Waltham, MA, USA), respectively. The sample was transported to a laboratory refrigerator and analyzed within 4 h.
As there was no established selective medium for K. salsicia isolation, a loop of each bottle of cheese brine solution was streaked onto nutrient agar (Sigma-Aldrich, St. Louis, MO, USA) and tryptone soya agar (Oxoid, Basingstoke, UK), and incubated for 24 h at 37°C, in triplicate. Additionally, the colonies were cultured on Columbia agar containing 5% sheep blood (bioMérieux, Marcy l’Etoile, France) after a 48 h incubation to detect non-hemolytic and lemon-yellow colonies—cultural characteristics of K. salsicia (Kandi et al., 2016; Sohn et al., 2015). Typical colonies (non-hemolytic and lemon-yellow colonies) were sub-cultured on nutrient agar. Because of the lack of proper guidelines for Kocuria spp., we used the positive control (PC) for Staphylococcus spp. mentioned by Sohn et al. (2015). Staphylococcus aureus American Type Culture Collection (ATCC) 29213 was purchased from the ATCC (Manassas, VA, USA) and used as a PC strain (Bruins et al., 2007). S. aureus ATCC 29213 is a clinical isolate that has been applied for enteric and infectious disease research (Soni et al., 2015). Furthermore, Escherichia coli ATCC 8739, enterohemorrhagic E. coli (EHEC) ATCC 43894, and Listeria monocytogenes ATCC 51776 were purchased from the ATCC.
DNA extraction was performed using the NucliSENS easyMAG system (bioMérieux). Briefly, each bacterial colony was added to lysis buffer (1.0 mL) and left to stand for 20 min at 25°C. The mixture was centrifuged at 15,770×g for 3 min using MIKRO 200 centrifuge (Hettich, Tuttlingen, Germany), and 1 mL of the supernatant was transferred to a well of a plastic vessel with 50 μL of magnetic silica and subjected to automated magnetic bead separation. DNA was then resuspended in 75 μL of elution buffer.
K. salsicia strains were isolated from cheese brine and identified via 16S rRNA sequencing. The 27F and 1492R primers were used for polymerase chain reaction (PCR). PCR products were sequenced using the same primers and ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA). Sequencing was performed using an Applied Biosystems 3730XL DNA Analyzer obtained from Bionics (Seoul, Korea). Each 16S rRNA sequence was analyzed via the basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the National Center for Biotechnology Information 16S rRNA database to identify K.salsicia.
To analyze the growth curve at different temperatures (5°C, 15°C, 25°C, 30°C, 37°C, and 42°C), the cheese brine isolates and S. aureus ATCC 29213 were cultured in nutrient agar (Sigma-Aldrich) for 24 h at 37°C. NB (200 μL; Sigma-Aldrich) was filter-sterilized using a 0.2-μm syringe filter (Millipore, Bedford, MA, USA) and then mixed with each isolate at 0.5 McFarland turbidity. After mixing, 200 μL of each isolate was transferred to a 96-well plate (SPL Life Sciences, Pocheon, Korea). The growth curves were generated at 24 h intervals over 288 h at 5°C; 8 h intervals over 96 h at 15°C, 25°C, and 30°C; and 2 h intervals over 24 h at 37°C and 42°C by measuring the optical density (OD) at 595 nm using a Multiskan FC microplate reader (Thermo Fisher Scientific). The experiment was repeated in triplicate for each isolate.
To analyze the growth of K.salsicia in a halophilic environment, three isolates (KS3, KS6, and KS8) among nine K. salsicia isolates were selected based on their relatively high biofilm production. KS3, KS6, and KS8 were compared with S. aureus ATCC 29213, E. coli ATCC 8739, EHEC ATCC 43894, and L. monocytogenes ATCC 51776 at a salt concentration of approximately 15% (w/v) based on the salt concentration of cheese brine in farmstead cheese manufacturing and different temperatures (Bintsis and Papademas, 2002; Bruins et al., 2007; Haastrup et al., 2018; Ingham et al., 2000; Larson et al., 1999). These four foodborne pathogens were used as quality control strains for analyzing the growth curve at 15% salt concentration. NB (200 μL; Sigma-Aldrich) containing 15% NaCl (Sigma-Aldrich) was filter-sterilized using a 0.2-μm syringe filter (Millipore) and then mixed with each strain at 0.5 McFarland turbidity. After mixing, 200 μL of each strain was transferred to a 96-well plate (SPL Life Sciences). The growth curves were generated at 24 h intervals over 288 h at 5°C; 8 h intervals over 96 h at 15°C, 25°C, and 30°C; and 2 h intervals over 24 h at 37°C and 42°C by measuring the OD at 595 nm using a Multiskan FC microplate reader (Thermo Fisher Scientific). The experiment was repeated in triplicate for each strain.
The biofilm-forming ability of the K. salsicia isolates was evaluated as previously described (Jeong et al., 2018). In brief, each colony of the isolates was added to 200 μL of PBS (Sigma-Aldrich), NB (Sigma-Aldrich), and NB containing 15% NaCl and set to 0.5–0.6 McFarland turbidity. To assess the extent of biofilm formation in each microplate, 200 μL of each sample was transferred to a 96-well polystyrene culture plate and incubated at 5°C, 15°C, 25°C, 30°C, 37°C, and 42°C for 24 h. The culture medium was discarded, and the microplate was washed twice with 200 μL of PBS (Sigma-Aldrich). Cells from adherent biofilms were stained with 0.1% (w/v) crystal violet (100 μL; Sigma-Aldrich) for 15 min at room temperature (20°C–25°C) and rinsed twice with PBS (Sigma-Aldrich). After removing the dye from stained cells using 99% ethanol (200 μL), the amount of biofilm was quantified by measuring the absorbance of the solution at 595 nm using a Multiskan FC microplate reader (Thermo Fisher Scientific). The experiment was performed in triplicate.
Antimicrobial susceptibility tests were performed using the VITEK® 2 instrument (bioMérieux) with gram-positive susceptibility (AST-GP) cards (bioMérieux). The AST-GP cards contained amikacin (AMK), chloramphenicol (CHL), clindamycin (CLI), gentamicin (GEN), cefpodoxime (POD), enrofloxacin (ENO), erythromycin (ERY), marbofloxacin (MAR), minocycline (MIN), nitrofurantoin (NIT), pradofloxacin (PRA), and trimethoprim/sulfamethoxazole (SXT), and the AMK, CHL, CLI, GEN, ENO, ERY, MAR, MIN, NIT, PRA, and SXT results were interpreted in accordance with the Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals in the Clinical and Laboratory Standards Institute guidelines (CLSI VET01S; CLSI, 2020a). However, owing to the lack of POD minimum inhibitory concentration criteria in CLSI VET01S, Performance Standards for Antimicrobial Susceptibility Testing in the CLSI was used to interpret the result (CLSI, 2020b). As breakpoints for K.salsicia have not been established, the antimicrobial resistance test was performed with reference to criteria used for S. aureus ATCC 29213 (Sohn et al., 2015).
Data for biofilm formation and growth curve analysis are presented as the mean±SD. GraphPad Prism 7.00 (GraphPad Software, San Diego, CA, USA) was used for data analyses. Biofilm formation data were analyzed using ANOVA followed by the Tukey method. p<0.05 was considered significant.
Results and Discussion
We identified nine K. salsicia isolates (KS1, KS2, KS3, KS4, KS5, KS6, KS7, KS8, and KS9) from a farmstead cheese house in Korea. The salt concentration and pH of the brine were 15%–18% (w/v) and 5.3, respectively. All K. salsicia strains were from different agar plates. Hemolytic K. salsicia colonies were not observed on Columbia agar containing 5% sheep blood but hemolytic S. aureus ATCC 29213 colonies were recorded. Consistently, 16S rRNA sequencing confirmed taxa of the cheese brine isolates as K. salsicia at the species level and their sequences were submitted to GenBank under accession numbers MW301599 for K. salsicia 1 (KS1), MW301601 for K. salsicia 2 (KS2), MW301600 for K. salsicia 3 (KS3), MW301603 for K. salsicia 4 (KS4), MW301604 for K. salsicia 5 (KS5), MW301605 for K.salsicia 6 (KS6), MW301606 for K. salsicia 7 (KS7), MW301607 for K. salsicia 8 (KS8), and MW301608 for K. salsicia 9 (KS9).
The growth curves of KS1–KS9 were compared with that of S. aureus ATCC 29213 by measuring the OD of the cultures at 595 nm (Fig. 1). Growth of all K. salsicia isolates was observed at all temperatures (5°C, 15°C, 25°C, 30°C, 37°C, and 42°C), suggesting that it is a psychotropic bacterium. The growth of K. salsicia strains was the highest during the exponential phase at 5°C and 15°C, which lasted for at least 96 and 288 h, respectively (Figs. 1A, B). OD values of K. salsicia isolates (0.12–0.71) were higher than S. aureus ATCC 29213 (0.12–0.29) at 5°C (Fig. 1A). At 25°C and 30°C, K. salsicia isolates were in the stationary phase for approximately 60 h. All K. salsicia isolates showed OD values higher than those of S. aureus ATCC 29213 (PC; Figs. 1C, D).
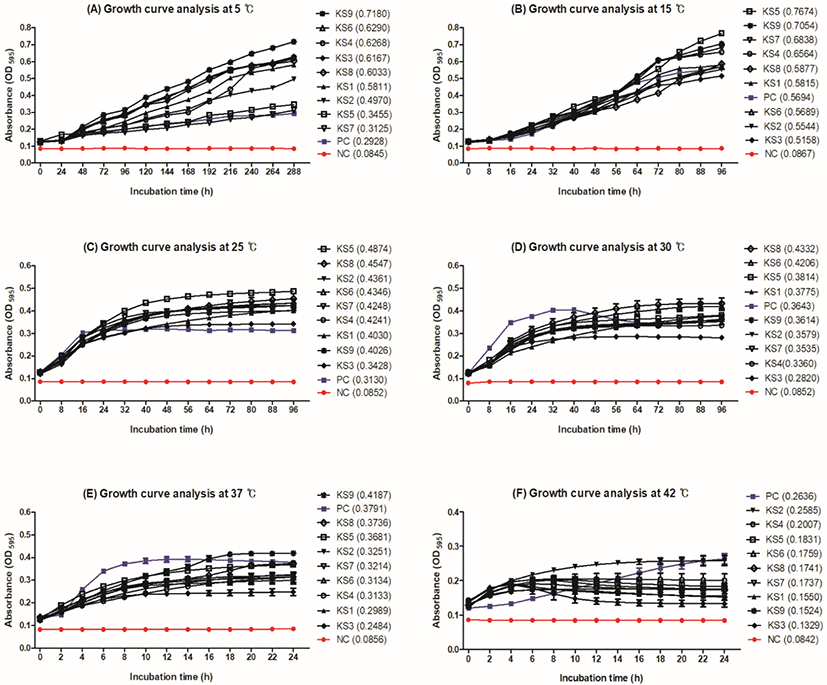
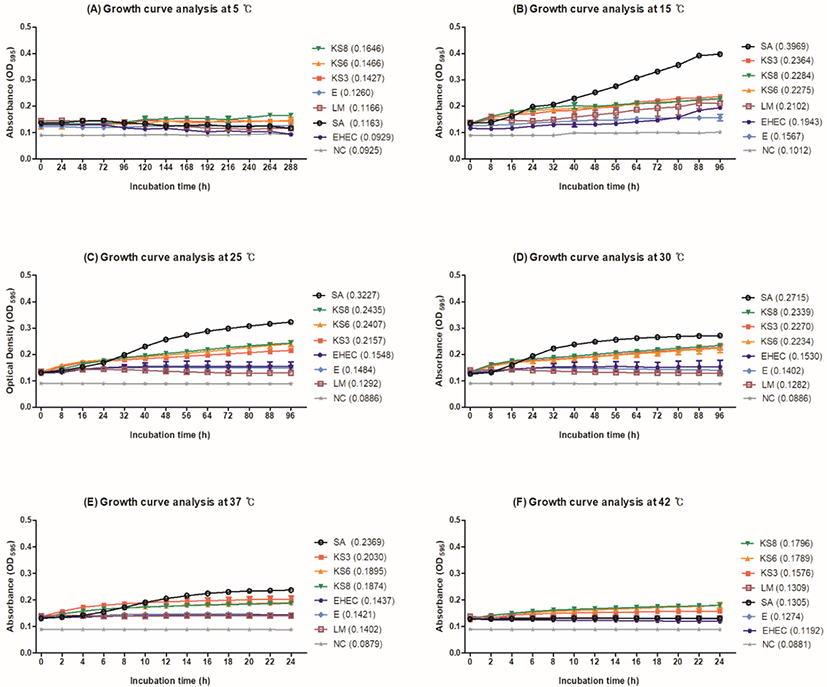
Growth curves of KS3, KS6, and KS8 were compared with those of S. aureus ATCC 29213, E. coli ATCC 8739, EHEC ATCC 43894, and L. monocytogenes ATCC 51776 by measuring the OD of the cultures at 595 nm (Fig. 2). All microorganisms grew at 15°C, 25°C, 30°C, 37°C, and 42°C in NB containing 15% NaCl. S. aureus ATCC 29213 showed higher growth than the other strains tested. Particularly, S. aureus ATCC 29213 showed the highest OD value of approximately 0.40 at 15°C (Fig. 2B). The growth of KS3, KS6, and KS8 increased gradually at 15°C, 25°C, 30°C, 37°C, and 42°C; KS6 and KS8 showed slow growth at 5°C (Fig. 2A). E. coli ATCC 8739, EHEC ATCC 43894, and L. monocytogenes ATCC 51776 showed lower OD values (approximately 0.1 to 0.2) than S. aureus ATCC 29213 and K. salsicia isolates at all temperatures. In general, salt processing promotes the syneresis of whey from the curd, thereby reducing the moisture content of the cheese (McMahon et al., 2009). Although cheese salting is believed to decrease the population of undesirable contaminants, brine can also serve as a reservoir for certain salt-tolerant pathogens (Bintsis and Papademas, 2002). The psychrotroph L. monocytogenes survives for longer periods in brines stored at 4°C than in those stored at 12°C (Larson et al., 1999). E. coli O157:H7 and Salmonella Typhimurium can survive for several weeks in brine (Ingham et al., 2000). Kocuria spp. grow at a temperature range of 4°C–43°C and tolerate up to 15% NaCl concentration (Kim et al., 2004). Consistent with these studies, our results indicated that K. salsicia strains grew at a temperature range of 5°C–42°C and tolerated 15% NaCl concentration (Fig. 2). Although cheese brine does not provide a favorable condition for microorganisms to grow owing to its high salt concentration, it can cause cross-contamination because of the whey derived from cheese and improper temperature control (Mehli et al., 2017).
The biofilm-forming ability of K. salsicia isolates (KS1–KS9) was evaluated in diverse temperatures and media. Although K. salsicia isolates did not grow well in PBS, the OD595 value of KS9 (0.065) was significantly higher than that of S. aureus ATCC 29213 (0.052) at 37°C (p<0.05; Table 1). Moreover, the biofilm-forming ability of KS6 was significantly different from that of S. aureus ATCC 29213 at the temperatures tested in NB (p<0.05; Table 2). The highest OD595 value for biofilms was observed for KS8 (0.371) followed by KS3 (0.361) at 30°C (p<0.05). Notably, KS3, KS6, and KS8 showed better biofilm production abilities than S. aureus ATCC 29213 at four of the tested temperatures (25°C, 30°C, 37°C, and 42°C; p<0.05). KS6 showed a significant difference in biofilm formation compared with S. aureus ATCC 29213, even at 5°C (p<0.05). In NB containing 15% NaCl, all microorganisms grew at 5°C, 15°C, 25°C, 30°C, 37°C, and 42°C and formed biofilm. Among temperatures, all K. salsicia isolates (KS1–KS9) showed higher OD values (range 0.054 to 0.066) than S. aureus ATCC 29213 at 15°C (p<0.05; Table 3).
Most cases of Kocuria spp. infection have been hypothesized to be associated with catheter-associated bloodstream infections; however, this association was not established until recently (Barnes et al., 1999; Sohn et al., 2015). Only a few studies have reported the biofilm-forming ability of Kocuria spp. Therefore, studying the biofilm-forming ability of K.salsicia may provide valuable insights into the biofilm-forming potential of other members of this genus (Purty et al., 2013). Furthermore, temperature is a key regulator of bacterial biofilm formation, and the temperature changes occurring in food, as well as hospital conditions, influence biofilm formation by microorganisms (Di Ciccio et al., 2015; Nilsson et al., 2011).
Consistent with the findings of the present study, another study reported that the effect of the growth temperature on the formation of S. aureus biofilm is influenced by several environmental factors such as nutrient availability and growth medium (Banks and Bryers, 1991; de Jesus Pimentel-Filho et al., 2014). In the present study, the OD values of K. salsicia KS1–KS9 and S. aureus ATCC 29213 were different; however, the patterns of OD values in all temperatures and media were similar. As brine containing cheese whey may have abundant nutrients for the growth of microorganisms, it is important to evaluate the biofilm formation ability of K. salsicia isolates in different nutrient sources. KS3, KS6, and KS8 showed higher growth rates than S. aureus ATCC 29213; this difference in growth rates was larger at 25°C and 30°C than that at 37°C and 42°C. Further, low temperatures increase the hydrophilic properties of cells and alter the ability of the bacteria to adhere to hydrophobic materials such as polystyrene and the effect of temperature on biofilm formation also depends on the presence or absence of NaCl (Rode et al., 2007). Auto-aggregation of microorganisms increases with increased NaCl concentration; this auto-aggregation is highly correlated with the biofilm formation by foodborne pathogens (Xu et al., 2010). To the best of our knowledge, biofilm formation under various temperatures, nutrient conditions, and environmental conditions has not been extensively investigated for Kocuria spp. Our results suggest that K. salsicia can grow at a wide range of temperatures and survive in the cheese brine tank. Altuntas et al. (2004) reported that K. rosea, a catheter-associated bacterium, is vancomycin sensitive; however, antimicrobial treatment is ineffective until the catheter is removed, indicating that the biofilm formation on the surface of the catheter can protect the bacterial community from antimicrobial action (Savini et al., 2010). Thus, the ability to form biofilms can facilitate the co-existence of halophilic bacteria in the biofilm in the brine, leading to the contamination of the final products and damage to equipment. It could also lead to the development of resistance to antibacterial agents or disinfectants, resulting in serious hygiene problems and economic losses (Barnes et al., 1999).
The antimicrobial susceptibility of the K. salsicia isolates obtained from cheese brine was investigated (Table 4). Five of the nine isolates (55.6%) were MDR, showing resistance to at least three different classes of antimicrobials. Moreover, the isolates showed higher resistance to the fluoroquinolone class of antimicrobials, including MAR, ENO, and PRA, than to other antimicrobials. Kocuria spp. are sensitive to ampicillin, CLI, ERY, GEN, SXT, and cotrimoxazole antimicrobials (Savini et al., 2010; Sohn et al., 2015). Becker et al. (2008) reported that quinolone antimicrobials are effective against Kocuria spp. However, in our results, the K. salsicia isolates showed high minimal inhibitory concentration values against fluoroquinolone antimicrobials ENO and MAR (Table 4). These results suggest that Kocuria spp. have developed antimicrobial resistance to quinolones over the past decade. The antimicrobials sold for use in Korean livestock farms have increased by more than 70%, from 57 tons in 2010 to 97 tons in 2019 (APQA, 2019). The sales of ENO account for more than 70%–80% of quinolone antimicrobials and more than 2 tons of MAR have been sold since 2014 (APQA, 2019). In the present study, most antimicrobials were effective against the K. salsicia isolates (KS1–KS9); however, five of the nine isolates showed resistance to at least three antimicrobials. A previous study reported that MDR bacteria isolated from the cheese-making environment mainly acquired resistance genes from the environment and animal facilities (Kang et al., 2018). As antimicrobial resistance genes can be transferred between bacteria, which may occur during food production, it is necessary to perform antimicrobial stewardship at the farming stage (Jang et al., 2020).
1) – indicates a negative result for antimicrobial resistance against more than three antimicrobial categories.
2) + indicates a positive result for antimicrobial resistance against more than three antimicrobial categories.
MIC, minimum inhibitory concentration; AMK, amikacin; CHL, chloramphenicol; CLI, clindamycin; GEN, gentamicin; POD, cefpodoxime; ENO, enrofloxacin; ERY, erythromycin; MAR, marbofloxacin; MIN, minocycline; NIT, nitrofurantoin; PRA, pradofloxacin; SXT, trimethoprim/ sulfamethoxazole; (R), resistant; (I), intermediate; (S), susceptible; MDR, multidrug-resistant; PC, positive control (Staphylococcus aureus ATCC 29213); KS1–KS9, K. salsicia KS1–KS9; ATCC, American Type Culture Collection.
Conclusion
Nine K. salsicia strains (KS1–KS9) were for the first time isolated from cheese brine in this study. They grew at a wide range of temperatures, had potential biofilm-forming ability, and showed antimicrobial resistance. The results of the current study showed that brine could serve as an important reservoir for various halotolerant or halophilic microorganisms. Therefore, careful monitoring and hygienic handling of cheese brine are needed to prevent microbial contamination of the final product during cheese production in farmstead dairy plants. A major limitation of this study is that we did not evaluate the halophile-related gene clusters using whole genome sequencing nor perform a co-incubation growth analysis using cheese starter strains. Thus, further investigation is warranted to assess the halophilic characteristics of K. salsicia isolated from cheese brine and fate of K. salsicia during the ripening process.













